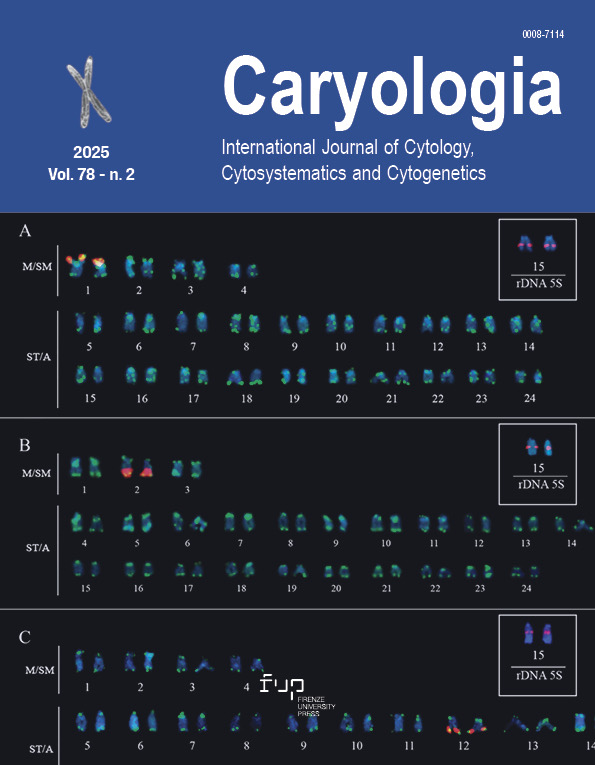
Karyotype analysis and chromosome evolution in Menyanthaceae using FISH
DOI:
https://doi.org/10.36253/caryologia-3431Keywords:
Menyanthaceae, karyotype analsis, Fluorescence in situ Hybridization (FISH), 45S rDNA, 5S rDNA, polyploidyAbstract
The Menyanthaceae, an aquatic plant family, is distinguished by extensive polyploidy and heterostyly. This study marks the first cytogenetic characterization of four Menyanthaceae species from Korea – Menyanthes trifoliata, Nymphoides peltata, N. indica, and N. coreana – employing fluorescence in situ hybridization (FISH) with 45S and 5S rDNA probes. All four species exhibit exclusively metacentric chromosomes, with M. trifoliata and N. peltata being hexaploid (2n = 54), N. coreana tetraploid (2n = 36), and N. indica diploid (2n = 18). FISH mapping revealed between one to four 45S rDNA loci and one to three 5S rDNA loci per species, showing that rDNA site number does not correlate directly with ploidy level. The karyotypic data suggest a conserved base chromosome number (x = 9) and largely symmetrical karyotypes across these species. Notably, M. trifoliata presents fewer rDNA loci than expected for a hexaploid, indicating genomic rearrangements and rDNA locus loss through diploidization. These observations highlight an evolutionarily stable genome structure in M. trifoliata, despite its polyploid nature. This study elucidates the chromosomal organization and evolutionary dynamics of the Menyanthaceae, emphasizing the role of polyploidy and rDNA evolution in genome structuring. The findings enhance our understanding of plant cytogenetics in aquatic ecosystems and serve as a foundation for further comparative genomic and evolutionary studies in Menyanthaceae.
Downloads
References
Abbo, S., Miller, T. E., Reader, S. M., Dunford, R. P. and King, I. P. 1994. Detection of ribosomal DNA sites in lentil and chickpea by fluorescent in situ hybridization. – Genome 37: 713–716.
Barrett, S. C. H. 1992. Heterostylous genetic polymorphisms: Model systems for evolutionary analysis. – Am. Nat. 139: 421–435.
Barrett, S. C. H. and Shore, J. S. 2008. New insights on heterostyly: Comparative biology, ecology and genetics. – Philos. Trans. R. Soc. B 363: 491–508.
Cook, C. D. K. 1996. Aquatic Plant Book. SPB Academic Publishing, Amsterdam.
Dulberger, R. and Ornduff, R. 2000. Stigma morphology in distylous and non-heterostylous species of Villarsia (Menyanthaceae). – Plant Syst. Evol. 225: 171–184.
Fultz, D. and Pikaard, C. S. 2023. Reaching for the off switch in nucleolar dominance. – Plant J. 115: 642–652.
Gillett, J. M. 1968. The systematics of the Asiatic and American populations of Fauria crista-galli (Menyanthaceae). – Can. J. Bot. 46: 92–96.
Haddadchi, A. 2013. Stylar polymorphism, reciprocity and incompatibility systems in Nymphoides montana (Menyanthaceae) endemic to southeastern Australia. – Plant Syst. Evol. 299: 389–401.
Haddadchi, A. and Fatemi, M. 2015. Self-compatibility and floral traits adapted for self-pollination allow homostylous Nymphoides geminata (Menyanthaceae) to persist in marginal habitats. – Plant Syst. Evol. 301: 239–250.
Ilnicki, T. 2014. Cytogenetic analysis of selected species using fluorescence in situ hybridization (FISH). – Caryologia 67: 192–196.
Kadereit, J. W. 2007. Menyanthaceae. In: Kadereit, J.W. and Jeffrey, C. (eds.), The Families and Genera of Vascular Plants, vol. 8. Springer, Berlin, pp. 225–242.
Kirov, I., Divashuk, M., Van Laere, K., Soloviev, A., and Khrustaleva, L. 2014. An easy “SteamDrop” method for high quality plant chromosome preparation. – Molecular Cytogenetics. 7: 1–10.
Kovarik, A., Dadejova, M., Lim, Y. K., Chase, M. W., Clarkson, J. J., Knapp, S. and Leitch, A. R. 2008. Evolution of rDNA in Nicotiana allopolyploids: a potential link between rDNA homogenization and epigenetics. – Ann. Bot. 101: 815–823.
Leitch, I. J. and Leitch, A. R. 2013. Genome size diversity and evolution in land plants. – New Phytol. 200: 3–18.
Li, L. J. and Arumuganathan, K. 2001. Physical mapping of sorted chromosomes 45S and 5S rDNA on maize metaphase by FISH. – Hereditas 134: 141–145.
Martel, E., Poncet, V., Lamy, F., Siljak-Yakovlev, S., Lejeune, B. and Sarr, A. 2004. Chromosome evolution of Pennisetum species (Poaceae): implications of ITS phylogeny. – Plant Syst. Evol. 249: 139–149.
Mlinarec, J., Malenica, N., Garfí, G., Bogunić, F., Satovic, Z., Siljak-Yakovlev, S. and Besendorfer, V. 2012. Evolution of polyploid Anemone was accompanied by rDNA locus loss. – Ann. Bot. 110: 703–712.
Ornduff, R. 1970. Cytogeography of Nymphoides (Menyanthaceae). – Taxon 19: 715–719.
Ornduff, R. 1987. Distyly and incompatibility in Villarsia congestiflora (Menyanthaceae), with comparative remarks on V. capitata. – Plant Syst. Evol. 159: 81–83.
Ornduff, R. 1992. Intrapopulation variation in the breeding system of Villarsia lasiosperma (Menyanthaceae). Plant Syst. Evol. 180: 227–233.
Peruzzi, L. and Cesca, G. 2004. Chromosome numbers of flowering plants from Calabria, S. Italy, II. – Informatore Bot. Ital. 36: 79–85.
Raabová, J., Münzbergová, Z. and Fischer, M. 2010. Consequences of polyploidization for phenotypic variation, fitness, and mating patterns in diploid and hexaploid Anemone nemorosa. – Ann. Bot. 105: 527–540.
Rosato, M., Kovarik, A., Garilleti, R. and Rosselló, J. A. 2015. Evolutionary site-number changes of ribosomal DNA loci during speciation. – AOB Plants 7: plv135.
Semple, J. C. and Watanabe, K. 2023. Index to chromosome numbers in Asteraceae (ICNA). – Plant Cytogenet. 58: 289–312.
Silvestri, M. C., Ortiz, A. M. and Lavia, G. I. 2015. rDNA loci and heterochromatin positions support a distinct genome type for ‘x = 9 species’ of section Arachis (Leguminosae). –Plant Syst. Evol. 301: 555–562.
Soltis, D. E. and Soltis, P. S. 2016. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. – Plant Biol. 30: 159–165.
Stace, C. A. 2000. Cytology and cytogenetics as a fundamental taxonomic resource for the 20th and 21st centuries. – Taxon 49: 451–477.
Stebbins, G. L. 1971. Chromosomal Evolution in Higher Plants. Edward Arnold, London.
Tippery, N. P., Les, D. H. and Hilu, K. W. 2008. Phylogeny and character evolution in the aquatic plant family Menyanthaceae. – Syst. Bot. 33: 736–751.
Tippery, N. P., Les, D. H. and Schafer, J. L. 2009. Phylogenetics of Nymphoides (Menyanthaceae): Evidence for a polyphyletic origin of aquatic heterophylly. – Syst. Bot. 34: 595–612.
Watanabe, K. 2022. Chromosomal evolution and descending dysploidy in Menyanthaceae. – Plant Genome Evol. 9: 221–235.
Weiss-Schneeweiss, H., Schneeweiss, G. M., Stuessy, T. F. and Mabuchi, T. 2013. Evolutionary patterns of rDNA loci in polyploid plants. – Ann. Bot. 111: 457–468.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Hye-rin Kim, Kweon Heo

This work is licensed under a Creative Commons Attribution 4.0 International License.
- Copyright on any open access article in a journal published byCaryologia is retained by the author(s).
- Authors grant Caryologia a license to publish the article and identify itself as the original publisher.
- Authors also grant any third party the right to use the article freely as long as its integrity is maintained and its original authors, citation details and publisher are identified.
- The Creative Commons Attribution License 4.0 formalizes these and other terms and conditions of publishing articles.
- In accordance with our Open Data policy, the Creative Commons CC0 1.0 Public Domain Dedication waiver applies to all published data in Caryologia open access articles.