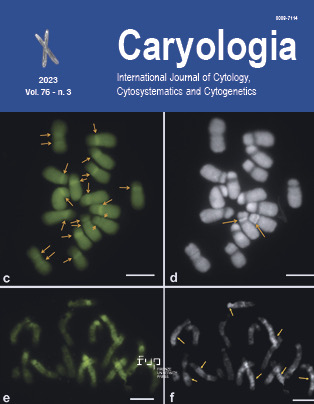
Chromosomal characterization mediated by karyomorphological analysis and differential banding pattern in fenugreek (Trigonella foenum-graecum L.): a neglected legume
DOI:
https://doi.org/10.36253/caryologia-2159Keywords:
Trigonella foenum-graecum, Karyotype, CMA-DAPI, AgNOR, FenugreekAbstract
Fenugreek or Trigonella foenum-graecum L. is a commercially important yet neglected crop of the family Fabaceae, with potent medicinal applications, and can treat several diseases as well. Conventional breeding studies for higher yields of commercial crops largely depend on chromosomal information of the particular species. Despite a number of cytological research being conducted on T. foenum-graecum, a complete characterization of its chromosomes has not been achieved due to the limitations of traditional karyotype analysis methods. A range of chromosomal markers are advantageous to characterize at full extent and identify individual chromosomes rather than relying on only physical metrics. Thus, in this study, in addition to giemsa staining, other approaches like fluorochrome and silver staining were used for the precise karyomorphological analysis of this species. Enzyme maceration and air drying (EMA) based fluorochrome banding with GC-specific stain Chromomycin A3 (CMA), and AT-specific stain 4’,6-diamidino-2-phenylindole (DAPI) applied for the first time for chromosome characterization. The results showed 2n = 16 chromosomes in metaphase cells, with karyotype formula of 2m+6sm. The unique banding pattern observed in the CMA/DAPI and AgNOR staining highlights the AT and GC-rich regions as well as the nucleolar organizer regions (NORs). All this crucial information can further assist in conducting breeding studies of more precision with simultaneously encouraging similar studies that need to be done in other unexploited species of importance.
Downloads
References
Acharya SN, Thomas JE, Basu SK. 2008. Fenugreek, an alternative crop for semiarid regions of North America. Crop Sci 48:841–853. https://doi.org/10.2135/cropsci2007.09.0519
Agarwal K, Gupta PK. 1983. Cytological studies in the genus Trigonella Linn. Cytologia 48:771–779. https://doi.org/10.1508/cytologia.48.771
Ahmad F, Acharya SN, Mir Z, Mir PS. 1999. Localization and activity of rRNA genes on fenugreek (Trigonella foenum-graecum L.) chromosomes by fluorescent in situ hybridization and silver staining. Theor Appl Genet 98:179–185. https://doi.org/10.1007/s001220051056
Al-Jasass FM, Al-Jasser MS. 2012. Chemical Composition and Fatty Acid Content of Some Spices and Herbs under Saudi Arabia Conditions. Sci World J. 2012:1–5. https://doi.org/10.1100/2012/859892
Andras SC, Hartman TPV, Alexander J et al. 2000. Combined PI–DAPI staining (CPD) reveals NOR asymmetry and facilitates karyotyping of plant chromosomes. Chromosome Res. 8:387–391. https://doi.org/10.1023/A:1009258719052
Arya ID, Rao SR, Raina SN. 1988. Cytomorphological studies of Trigonella foenum-graecum autotetraploids in three (C1, C2, C3) generation. Cytologia 53:525–534. https://doi.org/10.1508/cytologia.53.525
Bairiganjan GC, Patnaik SN. 1989. Chromosomal evolution in Fabaceae. Cytologia 54:51–64.
Barros e Silva AE, Guerra M. 2010. The meaning of DAPI bands observed after C-banding and FISH procedures. Biotech Histochem 85:115–125. https://doi.org/10.3109/10520290903149596
Basu S. 2023. Elucidating karyotype structure and affinity through application of karyomorphological parameters and multivariate analysis, as discerned from the study of four important legumes. Nucleus 66:39–46. https://doi.org/10.1007/s13237-023-00416-8
Berjano R, Roa F, Talavera S, Guerra M. 2009. Cytotaxonomy of diploid and polyploid Aristolochia (Aristolochiaceae) species based on the distribution of CMA/DAPI bands and 5S and 45S rDNA sites. Plant Syst Evol 280:219–227. https://doi.org/10.1007/s00606-009-0184-6
Biddak L. 1996. Inter-and intraspecific chromosomal variations in four species of Trigonella L. J Union Arab Biol, Cairo 3:203–215.
Bloom SE, Goodpasture C. 1976. An improved technique for selective silver staining of nucleolar organizer regions in human chromosomes. Hum Genet 34:199–206. https://doi.org/10.1007/BF00278889
Das AB, Mohanty S, Das P. 2001. Cytophotometric estimation of 4C DNA content and karyotype analysis in ten cultivars of Trigonella foenum-graecum. Iran J Bot 9:1–9.
Das AB, Mohanty S, Das P. 2002. Cytophotometric estimation of 4C DNA content and karyotype analysis in ten cultivars of Trigonella foenum-graecum-II. Iran J Bot 9:151–159.
Das AB, Mohanty S, Thangaraj T, Das P. 2000. Variation of 4C DNA content and karyotype in nine cultivars of fenugreek (Trigonella foenum-graecum L.). J Herbs Spices Med Plants 7:25–32. https://doi.org/10.1300/J044v07n01_04
de Melo NF, Guerra M. 2003. Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Ann Bot 92:309–316. https://doi.org/10.1093/aob/mcg138
de Moraes AP, dos Santos Soares Filho W, Guerra M. 2007. Karyotype diversity and the origin of grapefruit. Chromosome Res 15:115–121. https://doi.org/10.1007/s10577-006-1101-2
Dydak M, Kolano B, Nowak T, Siwinska D, Maluszynska J. 2009. Cytogenetic studies of three European species of Centaurea L. (Asteraceae). Hereditas 146:152–161. https://doi.org/10.1111/j.1601-5223.2009.02113.x
Feyzi S, Varidi M, Zare F, Varidi MJ. 2015. Fenugreek (Trigonella Foenum Graecum) Seed Protein Isolate: Extraction Optimization, Amino Acid Composition, Thermo and Functional Properties. J Sci Food Agric. 95:3165–3176. https://doi.org/10.1002/jsfa.7056
Guerra M. 2000. Patterns of heterochromatin distribution in plant chromosomes. Genet Mol Biol. 23:1029–1041. https://doi.org/10.1590/S1415-47572000000400049
Jahan B, Vahidy AA, Ali SI. 1994. Chromosome numbers in some taxa of Fabaceae mostly native to Pakistan. Ann Mo Bot Gard 792–799. https://doi.org/10.1508/cytologia.54.51
Jiménez R, Burgos M, de La Guardia RD. 1988. A study of the Ag-staining significance in mitotic NOR’s. Heredity 60:125–127. https://doi.org/10.1038/hdy.1988.18
Kar K, Sen S. 1991. A comparative karyological study of root and embryo tissue of a few genera of Leguminosae. Cytologia 56:403–408. https://doi.org/10.1508/cytologia.56.403
Kodama Y, Yoshida MC, Sasaki M. 1980. An improved silver staining technique for nucleolus organizer regions by using nylon cloth. Jpn J Hum Genet 25:229–233. https://doi.org/10.1007/BF01997700
Kolano B, Saracka K, Broda-Cnota A, Maluszynska J. 2013. Localization of ribosomal DNA and CMA3/DAPI heterochromatin in cultivated and wild Amaranthus species. Sci Hortic 164:249–255. https://doi.org/10.1016/j.scienta.2013.09.016
Ladizinsky G, Vosa CG. 1986. Karyotype and C-banding in Trigonella section Foenumgraecum (Fabaceae). Plant Syst Evol 153:1–5. https://doi.org/10.1007/BF00989412
Laxmi V, Gupta MN, Datta SK. 1983. Investigations on an induced green seed coat colour mutant of Trigonella foenum-graecum L. Cytologia 48:373–378. https://doi.org/10.1508/cytologia.48.373
Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas 52:201–220. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x
Levin DA. 2002. The role of chromosomal change in plant evolution. Oxford University Press, New York, USA.
Maragheh FP, Janus D, Senderowicz M, Haliloglu K, Kolano, B. 2019. Karyotype analysis of eight cultivated Allium species. J Appl Genet 60:1–11. https://doi.org/10.1007/s13353-018-0474-1
Marcon AB, Barros ICL, Guerra M. 2005. Variation in chromosome numbers, CMA bands and 45S rDNA sites in species of Selaginella (Pteridophyta). Ann Bot 95:271–276. https://doi.org/10.1093/aob/mci022
Martin E, Akan H, Ekici M, Aytac Z. 2011. Karyotype analyses of ten sections of Trigonella (Fabaceae). Comp Cytogenet 5:105–121. https://doi.org/10.3897/compcytogen.v5i2.969
Melters DP, Bradnam KR, Young HA et al. 2013. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol 14:1–20. https://doi.org/10.1186/gb-2013-14-1-r10
Mikić A. 2015. Brief but alarming reminder about the need for reintroducing ‘Greek hay’ (Trigonella foenum-graecum L.) in Mediterranean agricultures. Genet Resour Crop Evol 62:951–958. https://doi.org/10.1007/s10722-015-0260-4
Mondin M, Aguiar-Perecin ML. 2011. Heterochromatin patterns and ribosomal DNA loci distribution in diploid and polyploid Crotalaria species (Leguminosae, Papilionoideae), and inferences on karyotype evolution. Genome 54:718–726. https://doi.org/10.1139/g11-034
Najafi S, Anakhatoon EZ, Birsin MA. 2013. Karyotype Characterisation of Reputed Variety of Fenugreek (Trigonella foenum-graecum) in West Azerbaijan-Iran. J Appl Biol Sci 7:23–26.
Petropoulos GA. 2002. Fenugreek: The Genus Trigonella. CRC Press, Boca Raton, Florida, USA.
Ranjbar M, Zahra H. 2016. Chromosome numbers and biogeography of the genus Trigonella (Fabaceae). Caryologia 69:223–234. https://doi.org/10.1080/00087114.2016.1169090
Rasheed MSAA, Wankhade MV, Saifuddin MSSK, Sudarshan MAR. 2015. Physico-chemical properties of fenugreek (Trigonella foenum-graceum L.) seeds. Int J Eng Res 4:68–70.
Santra I, Halder T, Ghosh B. 2021. Somatic and gametic chromosomal characterization with fluorescence banding of Giloy (Tinospora cordifolia): A berberine synthesizing important medicinal plant of India. Caryologia. 74:63–73. https://doi.org/10.36253/caryologia-1014
Santra I, Haque SM, Ghosh B. 2020. Giemsa C-banding Karyotype and Detection of Polymorphic Constitutive Heterochromatin in Nigella sativa L. Cytologia. 85:85–90. https://doi.org/10.1508/cytologia.85.85
Schweizer D. 1976. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma. 58:307–324. https://doi.org/10.1007/BF00292840
Shabir PA, Wani AA, Nawchoo IA. 2017. Banding Techniques in Chromosome Analysis. In: Bhat T, Wani A (eds) Chromosome Structure and Aberrations. Springer, New Delhi, pp 167–180 https://doi.org/10.1007/978-81-322-3673-3_8
Sharghi H, Azizi M, Moazzeni H. 2020. A karyological study of some endemic Trigonella species (Fabaceae) in Iran. Caryologia. 73:155–161. https://doi.org/10.13128/caryologia-184
She CW, Jiang XH. 2015. Karyotype analysis of Lablab purpureus (L.) sweet using fluorochrome banding and fluorescence in situ hybridization with rDNA probes. Czech J Genet Plant Breed. 51:110–116. https://doi.org/10.17221/32/2015-CJGPB
Stebbins GL. 1971. Chromosomal Evolution in Higher Plants. Edward Arnold Ltd., London.
Syed QA, Rashid Z, Ahmad MH, Shukat R, Ishaq A, Muhammad N, Rahman HUU. 2020. Nutritional and therapeutic properties of fenugreek (Trigonella foenum-graecum): a review. Int J Food Prop. 23:1777–1791. https://doi.org/10.1080/10942912.2020.1825482
Wani SA, Kumar P. 2018. Fenugreek: A Review on Its Nutraceutical Properties and Utilization in Various Food Products. J Saudi Soc Agri Sci. 17:97–106. https://doi.org/10.1016/j.jssas.2016.01.007
Yamamoto M. 2012. Recent progress on studies of chromosome observation in deciduous fruit trees. J Jpn Soc Hortic Sci. 81:305–313. https://doi.org/10.2503/jjshs1.81.305
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Indranil Santra, Diptesh Biswas, Biswajit Ghosh

This work is licensed under a Creative Commons Attribution 4.0 International License.
- Copyright on any open access article in a journal published byCaryologia is retained by the author(s).
- Authors grant Caryologia a license to publish the article and identify itself as the original publisher.
- Authors also grant any third party the right to use the article freely as long as its integrity is maintained and its original authors, citation details and publisher are identified.
- The Creative Commons Attribution License 4.0 formalizes these and other terms and conditions of publishing articles.
- In accordance with our Open Data policy, the Creative Commons CC0 1.0 Public Domain Dedication waiver applies to all published data in Caryologia open access articles.