Unfolding chromosomal uniqueness of the Scilloid ornamental Albuca virens by application of EMA based Giemsa- DAPI staining
DOI:
https://doi.org/10.36253/caryologia-2124Keywords:
Albuca virens, EMA, DAPI, meiosis, dysploidy, NORsAbstract
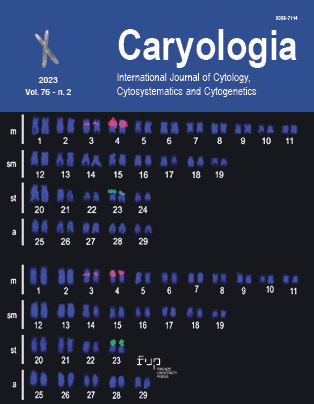
The cytogenetic features in the tribe Ornithogaleae of subfamily Scilloideae is a prerequisite for understanding genome evolution. Unfortunately, genomic or the foundational chromosomal features are neglected within majority of Ornithogaloids, incuding Albuca, one of the largest genus of the tribe. Albuca virens (Lindl.) J.C. Manning & Goldblatt is the only ornamental species of Albuca found to be exotic to India. Analysis of karyotype by EMA method followed by Giemsa and DAPI staining is the first step towards molecular cytogenetics attempted in this species and has currently brought significant resolution in chromosome morphology (especially NORs). The Indian population of A. virens with 2n=6 chromosomes, symmetric karyotype and heteromorphy of NORs provide excellent scope to navigate questions on dysploid origin of Albuca. The regular meiotic stages advocate genomic stability despite vegetative propagation and polysomaty in root cells. The comparative review of chromosomal evolution within Albuca has been discussed in relation to the Indian A. virens as a prototype.
Downloads
References
Angiosperm Phylogeny Group. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161: 105–121.
Angiosperm Phylogeny Group. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181: 1-20.
Arano H. 1963. Cytological studies in subfamily Carduoideae (Compositae) of Japan IX. Bot Mag Tokyo. 76: 32.
Bhattacharya S, Ghosh B, Choudhury M. 2016. A Simple Reliable Protocol for Cytogentically Stable Mass Propagation of Ornithogalum virens Lindl. Plant Tissue Cult Biotech. 26: 1-14.
Bhowmick BK, Jha S. 2022. A critical review on cytogenetics of Cucurbitaceae with updates on Indian taxa. Comp Cytogenet. 16: 93-126.
Bhowmick BK, Jha S. 2015. Differential heterochromatin distribution, flow cytometric genome size and meiotic behavior of chromosomes in three Cucurbitaceae species. Sci Hortic. 193: 322-329.
Bhowmick BK, Jha S. 2019. Differences in karyotype and fluorochrome banding patterns among variations of Trichosanthes cucumerina with different fruit size. Cytologia. 84: 237-245.
Bhowmick BK, Jha S. 2021. A comparative account of fluorescent banding pattern in the karyotypes of two Indian Luffa species. Cytologia. 86: 35-39.
Castiglione MR, Cremonini R. 2012. A fascinating island: 2n= 4. Plant Biosyst. 146: 711-726.
Fukui K. 1996. Plant chromosomes at mitosis. In: Fukui K, Nakayama S, editors. Plant chromosomes. Laboratory methods. Boca Raton, Tokyo: CRC Press, p. 1-18.
Fukui K, Iijima K. 1991. Somatic chromosome map of rice by imaging methods. Theor Appl Genet. 81: 589-596.
Greilhuber J, Speta F. 1976. C-banded karyotypes in the Scilla hohenackeri group, S.persica and Puschkinia (Liliaceae). Plant Syst Evol. 126: 149–188.
Griesbach RJ, Meyer F, Koopowitz H. 1990. Interspecific hybridization of Ornithogalum. Hort Science. 25: 1142d-1142.
Griesbach RJ, Meyer F, Koopowitz H. 1993. Creation of new flower colors in Ornithogalum via interspecific hybridization. J Am Soc Hortic Sci. 118: 409-414.
Goldblatt P, Manning JC. 2011. A review of chromosome cytology in Hyacinthaceae subfamily Ornithogaloideae (Albuca, Dipcadi, Ornithogalum and Pseudogaltonia) in sub-Saharan Africa. South Afr J Bot. 77: 581-591.
Guerra M, Santos KGBD, E Silva AEB, Ehrendorfer F. 2000. Heterochromatin banding patterns in Rutaceae-Aurantioideae—a case of parallel chromosomal evolution. Am J Bot. 87(5): 735–747.
Huziwara Y. 1962. Karyotype analysis in some genera of Compositae. VIII. Further studies on the chromosomes of Aster. Am J Bot. 49: 116–119.
Jha TB. 2021. Karyotype diversity in cultivated and wild Indian rice through EMA-based chromosome analysis. J Genet. 100: 81.
Jha TB, Saha PS, Jha S. 2020. A comparative karyo-morphometric analysis of Indian landraces of Sesamum indicum using EMA-giemsa and fluorochrome banding. Caryologia. 73: 81-88.
Jha TB, Bhowmick BK. 2021. Conservation of floral, fruit and chromosomal diversity: a review on diploid and polyploid Capsicum annuum complex in India. Mol Biol. 48: 5587-5605.
Johnson MA. 1999. Two new chromosome numbers in an unusual Albuca (Hyacinthaceae) from Saudi Arabia. Kew Bulletin. 437-444.
Jong K. 1991. Cytological observations in Albuca, including a survey of polymorphic variation in thesat-chromosome pair. Edinb J Bot. 48: 107–128.
Knudtzon SH, Stedje B. 1986. Taxonomy and cytology of the genus Albuca (Hyacinthaceae) in East Africa. Nord J Bot. 6: 773-786.
Kurata N, Omura T. 1978. Karyotype analysis in rice. 1. A new method for identifying all chromosome pairs. Japan J Genet. 53:251–255.
Lavania UC, Srivastava S. 1992. A simple parameter of dispersion index that serves as an adjunct to karyotype asymmetry. J Biosci. 17: 179–182.
Liang G, Chen H. 2015. Scaling chromosomes for an evolutionary karyotype: a chromosomal trade off between size and number across woody species. PLoS One. 10: e0144669.
Manning JC. 2020. Systematics of Ledebouria sect. Resnova (Hyacinthaceae: Scilloideae: Massonieae), with a new subtribal classification of Massonieae. S Afr J Bot. 133: 98-110.
Manning JC, Goldblatt P, Williamson G. 2009a. Hyacinthaceae. Pseudogaltonia liliiflora (Ornithogaloideae), a new species from the Richtersveld, Northern Cape. Bothalia. 39: 229–231.
Manning JC, Forest F, Fay MF, Devey S, Goldblatt P. 2009b. A molecular phylogeny and a revised classification of Ornithogaloideae (Hyacinthaceae) based on an analysis of four plastid DNA regions. Taxon. 58: 77–107.
Martinez-Azorin M, Crespo MB, Juan A, Fay MF. 2011. Molecular phylogenetics of subfamily Ornithogaloideae (Hyacinthaceae) based on nuclear and plastid DNA regions, including a new taxonomic arrangement. Ann Bot. 107: 1-37.
Martínez-Azorín M, Crespo MB, Alonso-Vargas MÁ, et al. 2023. Molecular phylogenetics of subfamily Urgineoideae (Hyacinthaceae): Toward a coherent generic circumscription informed by molecular, morphological, and distributional data. J Syst Evol. 61: 42-63.
Nath S, Jha TB, Mallick SK, Jha S. 2015. Karyological relationships in Indian species of Drimia based on fluorescent chromosome banding and nuclear DNA amount. Protoplasma. 252: 283-299.
Nath S, Sarkar S, Patil SD, et al. 2022. Cytogenetic Diversity in Scilloideae (Asparagaceae): a comprehensive recollection and exploration of karyo-evolutionary trends. Bot Rev. 1-43.
Öztürk D, Koyuncu O, Koray Yaylacı Ö, Özgişi K, Sezer O, Tokur S. 2014. Karyological studies on the four Ornithogalum L. (Asparagaceae) taxa from Eskişehir (Central Anatolia, Turkey). Caryologia. 67: 79-85.
Paszko B. 2006. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol. 258: 39-48.
Pedrosa A, Jantsch MF, Moscone EA, Ambros PF, Schweizer D. 2001. Characterisation of pericentromeric and sticky intercalary heterochromatin in Ornithogalum longibracteatum (Hyacinthaceae). Chromosoma. 110: 203–213.
Peruzzi L, Eroğlu HE. 2013. Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogenet. 7: 1–9.
Ravindran PN. 1977. Homologous Association in Somatic Cells of Ornithogalum virens. Cytologia. 42: 1-4.
Schweizer D. 1976. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma. 58: 307–324.
Seijo JG, Fernández A. 2003. Karyotype analysis and chromosome evolution in South American species of Lathyrus (Leguminosae). Am J Bot. 90: 980–987.
Sharma AK, Sharma A. 1956. Fixity in chromosome number of plants. Nature. 177: 335-336.
Stebbins GL. 1971. Chromosomal changes, genetic recombination and speciation. In: Barrington EJW, Willis AJ, editors. Chromosomal Evolution in Higher Plants. Edward Arnold Publishers Pvt. Ltd., London. p. 72–123.
Stedje B. 1989. Chromosome evolution within the Ornithogalum tenuifolium complex (Hyacinthaceae), with species emphasis on the evolution of bimodal karyotypes. Plant Syst Evol. 166: 79–89.
Stedje B. 1996. Karyotypes of some species of Hyacinthaceae from Ethiopia and Kenya. Nord J Bot. 16: 121–126.
Watanabe K, Yahara T, Denda T, Kosuge K. 1999. Chromosomal evolution in the genus Brachyscome (Asteraceae, Astereae): statistical tests regarding correlation between
changes in karyotype and habit using phylogenetic information. J Plant Res. 112: 145–161.
Yamamoto M, Takada N, Hirabayashi T, Kubo T, Tominaga S. 2010. Fluorescent staining analysis of chromosomes in pear (Pyrus spp.). J Jpn Soc Hortic Sci. 79: 23-26.
Yamamoto M, Terakami S, Yamamoto T. 2015. Enzymatic maceration/air-drying method for chromosome observations in the young leaf of pear (Pyrus spp.). Chromosome Sci. 18: 29-32.
Zarco CR. 1986. A new method for estimating karyotype asymmetry. Taxon. 35: 526–530.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 biplabkumar bhowmick, Sayani Nag

This work is licensed under a Creative Commons Attribution 4.0 International License.
- Copyright on any open access article in a journal published byCaryologia is retained by the author(s).
- Authors grant Caryologia a license to publish the article and identify itself as the original publisher.
- Authors also grant any third party the right to use the article freely as long as its integrity is maintained and its original authors, citation details and publisher are identified.
- The Creative Commons Attribution License 4.0 formalizes these and other terms and conditions of publishing articles.
- In accordance with our Open Data policy, the Creative Commons CC0 1.0 Public Domain Dedication waiver applies to all published data in Caryologia open access articles.