What defines a bimodal karyotype? Bimodality revisited
DOI:
https://doi.org/10.36253/caryologia-3034Keywords:
Avdulov, bimodal karyotype, chromosome asymmetry, chromosome variation, cytogenetics, StebbinsAbstract
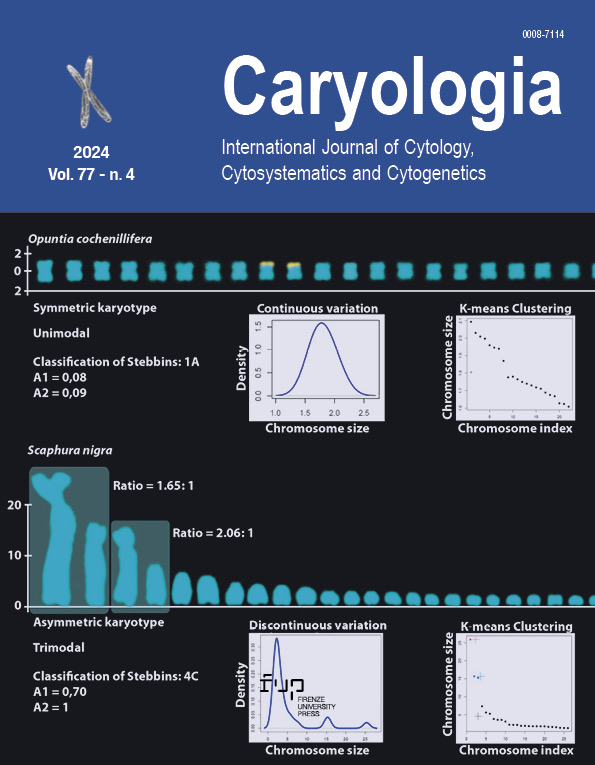
Bimodal karyotypes, initially defined by Avdulov, are characterized by one large and one small set of chromosomes, reflecting a particular type of karyotype asymmetry. Despite later discussions by Stebbins, the absence of a quantitative criterion has led to subjective classifications. This study revisits the concept of bimodality through a literature review and proposes an objective criterion based on the ratio between the smallest chromosome of the larger set and the largest of the smaller set. Chromosome morphology and asymmetry were analyzed in 32 species previously classified as bimodal. Statistical tests were applied to detect size discontinuities and assess bimodality. We propose two forms of bimodality, interchromosomal and intrachromosomal, considering differences in size and morphology. Our results show that Drosophila melanogaster and Scaphura nigra exhibit trimodal karyotypes. A ratio of ≥1.5:1 between chromosomal subsets provides a clear and objective criterion for defining bimodality, aligning with the original concepts of Avdulov and Stebbins.
Downloads
References
Alves LIF, Lima SAA, Felix LP. 2011. Chromosome characterization and variability in some Iridaceae from Northeastern Brazil. Genet Mol Biol 34(2):259-267. https://doi.org/10.1590/S1415-47572011000200016
Assis FNM, Souza BCQ, Medeiros-Neto E, Pinheiro F, Silva AEB, Felix LP. 2013. Karyology of the genus Epidendrum (Orchidaceae: Laeliinae) with emphasis on subgenus Amphiglottium and chromosome number variability in Epidendrum secundum. Bot J Linn Soc 172(3):329-344. https://doi.org/10.1111/boj.12045
Avdulov NP. 1931. Karyosystematische Untersuchung der Familie Gramineen. Bull Appl Bot Genet Plant Breed Suppl 44: 1-428.
Baeza M, Ruiz E, Negritto M. 2010. Comparative analysis in the Alstroemeria hookeri Lodd. (Alstroemeriaceae) complex Sensu Bayer (1987). Genet Mol Biol 33(1): 119-124. https://doi.org/10.1590/S1415-47572010005000012
Báez M, Vaio M, Dreissig S, Schubert V, Houben A, Pedrosa-Harand A. 2019. Together but different: The subgenomes of the bimodal Eleutherine karyotypes are differentially organized. Front Plant Sci 10:1170. https://doi.org/10.3389/fpls.2019.01170
Balslev H. 1996. Juncaceae. Fl Neotrop 68: 1-167. https://www.jstor.org/stable/4393863
Bennett MD, Smith JB, Seal AG. 1986. The karyotype of the grass Zingeria biebersteiniana (2n = 4) by light and electron microscopy. Can J Genet Cytol 28(4): 554-562. https://doi.org/10.1139/g86-081
Bennett ST, Kenton AY, Bennett MD. 1992. Genomic in situ hybridization reveals the allopolyploid nature of Milium montianum (Gramineae). Chromosoma 101: 420-424. https://doi.org/10.1007/BF00582836
Bertollo LAC, Takahashi CS, Moreira‐Filho O. 1983. Multiple sex chromosomes in the genus Hoplias (pisces: Erythrinidae). Cytologia 48:1-12. DOI: https://doi.org/10.1508/cytologia.48.1
Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LET. 2014. The quantitative architecture of centromeric chromatin. eLife 3:e02137. https://doi.org/10.7554/eLife.02137
Brito RO, Affonso PRAM, Silva Jr JC. 2010. Chromosomal diversity and phylogenetic inferences concerning thrips (Insecta, Thysanoptera) in a semi-arid region of Brazil. Genet Mol Res 9(4): 2230-2238.
Carta A, Bedini G, Peruzzi L. 2018. Unscrambling phylogenetic effects and ecological determinants of chromosome number in major angiosperm clades. Sci Rep 8(1): 1-14. https://doi.org/10.1038/s41598-018-32515-x
Castro JP, Medeiros-Neto E, Souza G, Alves LI, Batista FR, Felix LP. 2016. CMA band variability and physical mapping of 5S and 45S rDNA sites in Brazilian Cactaceae: Pereskioideae and Opuntioideae. Braz J Bot 39: 613-620. https://doi.org/10.1007/s40415-015-0248-5
Chase MW, Samuel R, Leitch AR, Guignard MS, Conran JG, Nollet F, Fletcher P, Jakob A, Cauz-Santos LA, Vignolle G, et al. 2023. Down, then up: non-parallel genome size changes and a descending chromosome series in a recent radiation of the Australian allotetraploid plant species, Nicotiana section Suaveolentes (Solanaceae). Ann Bot 131(1): 123–142. https://doi.org/10.1093/aob/mcac006
Chester M, Gallagher JP, Symonds VV, Silva AVC, Mavrodiev EV, Leitch AR, Soltis PS, Soltis DE. 2012. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). PNAS 109(4): 1176–1181. https://doi.org/10.1073/pnas.1112041109
Chiarini FE, Barboza GE. 2008. Karyological studies in Jaborosa (Solanaceae). Bot J Lin Soc 156(3):467-478. https://doi.org/10.1111/j.1095-8339.2007.00734.x
Crosland MWJ, Crozier RH. 1986. Myrmecia pilosula, an Ant with Only One Pair of Chromosomes. Science 231(4743): 1278. https://doi.org/10.1126/science.231.4743.1278.
de Azkue D, Martinez A. 1983. The chromosome complements of shrubby Oxalis species from South America. Plant Syst Evol 141: 187–197. https://doi.org/10.1007/BF00989001
Delaunay LN. 1923. Vergleichende karyologische Untersuchungen einiger Muscari Mill. und Bellevalia Lapeyr Arten. Monit Jard Bot Tbilisi 2(1): 24-55.
Goldblatt P, Snow N. 1991. Systematics and chromosome cytology of Eleutherine Herbert (Iridaceae). Ann Mo Bot Gard 78(4): 942–949. https://doi.org/10.2307/2399735
Greilhuber J. 1995. Chromosomes of the monocotyledons (general aspects). In: Randall PJ, Cribb PJ, Cutler DF, Humphries CJ. editors. Monocotyledons: systematics and evolution. Kew: Royal Botanic Gardens; p. 379–414.
Guerra M. 1986. Reviewing the chromosome nomenclature of Levan et al. Rev Bras Genet 9(4): 741–743.
Guerra M. 1988a. Introdução à citogenética geral. Rio de Janeiro: Guanabara koogan.
Guerra M. 1988b. Mitotic and meiotic analysis of a pericentric inversion associated with a tandem duplication in Eleutherine bulbosa. Chromosoma 97: 80-87. https://doi.org/10.1007/BF00331797
Guerra M. 2008. Chromosome numbers in plant cytotaxonomy: concepts and implications. Cytogenet Genome Res 120(3-4): 339–350. https://doi.org/10.1159/000121083
Guerra MS, Nogueira MTM. 1990. The cytotaxonomy of Emilia spp. (Asteraceae: Senecioneae) occurring in Brazil. Plant Syst Evol 170(3-4): 229–236. http://www.jstor.org/stable/23674239
Guerra M, Ribeiro T, Felix L. 2019. Monocentric chromosomes in Juncus (Juncaceae) and implications for the chromosome evolution of the family. Bot J Linn Soc 191(4): 475-483. https://doi.org/10.1093/botlinnean/boz065
Hartigan JA, Hartigan PM. 1985. The Dip Test of unimodality. Ann Statist 13(1): 70-84. https://doi.org/10.1214/aos/1176346577
Ibiapino A, Báez M, García MA, Costea M, Stefanović S, Pedrosa‑Harand A. 2022. Karyotype asymmetry in Cuscuta L. subgenus Pachystigma reflects its repeat DNA composition. Chromosome Res 30(1):91-107. doi: https://doi.org/10.1007/s10577-021-09683-0.
Jain AK. 2010. Data clustering: 50 years beyond K-means. Pattern Recognit Lett 31(8): 651-666. https://doi.org/10.1016/j.patrec.2009.09.011
Khoshoo TN, Ahuja MR. 1962. The Karyotype in Welwitschia mirabilis. Nature 193: 356–357. https://doi.org/10.1038/193356a0
Leach CR, Houben A, Timmisa JN. 2004. The B chromosomes in Brachycome. Cytogenet Genome Res 106(2-4):199–209. https://doi.org/10.1159/000079288.
Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas 52(2): 201-220. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x
Lisachov AP, Tishakova KV, Romanenko SA, Molodtseva AS, Prokopov DY, Pereira JC, Ferguson-Smith MA, Borodin PM, Trifonov VA. 2021. Whole-chromosome fusions in the karyotype evolution of Sceloporus (Iguania, Reptilia) are more frequent in sex chromosomes than autosomes. Philos Trans R Soc Lond B Biol Sci 376(1833):20200099. https://doi.org/10.1098/rstb.2020.0099
Lysak MA, Cheung K, Kitschke M, Bureš P. 2007. Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol 145(2): 402–410. https://doi.org/10.1104/pp.107.104380
Mayrose I, Lysak MA. 2021. The evolution of chromosome numbers: mechanistic models and experimental approaches. Genome Biol Evol 13(2):evaa220. https://doi.org/10.1093/gbe/evaa220
McKain MR, Wickett N, Zhang Y, Ayyampalayam S, McCombie WR, Chase MW, Pires JC, dePamphilis CW, Leebens-Mack J. 2012. Phylogenomic analysis of transcriptome data elucidates co-occurrence of a paleopolyploid event and the origin of bimodal karyotypes in Agavoideae (Asparagaceae). Amer J Bot 99(2):397–406. https://doi.org/10.3732/ajb.1100537
Medeiros-Neto E, Nollet F, Moraes AP, Felix LP. 2017. Intrachromosomal karyotype asymmetry in Orchidaceae. Genet Mol Biol 40(3): 610–619. https://doi.org/10.1590/1678-4685-GMB-2016-0264
Mesa A, Fontanetti CS, Ferreira A. 2010. The chromosomes and the sex determining mechanism of Scaphura nigra (Orthoptera, Ensifera, Tettigoniidae, Phaneropterinae). J Orthoptera Res19(2):239-242. http://hdl.handle.net/11449/19826
Moraes AP, Guerra M. 2010. Cytological differentiation between the two subgenomes of the tetraploid Emilia fosbergii Nicolson and its relationship with E. sonchifolia (L.) DC. (Asteraceae). Plant Syst Evol 287:113–118. https://doi.org/10.1007/s00606-010-0302-5
Moraes AP, Koehler S, Cabral JS, Gomes SS, Viccini LF, Barros F, Felix LP, Guerra M, Forni-Martins ER. 2017. Karyotype diversity and genome size variation in Neotropical Maxillariinae orchids. Plant Biol 19(2):298-308. https://doi.org/10.1111/plb.12527.
Nielsen MK, Wang J, Davis R, Bellaw JL, Lyons ET, Lear TL, Goday C. 2014. Parascaris univalens—a victim of large-scale misidentification? Parasitol Res 113(12): 4485–4490. https://doi.org/10.1007/s00436-014-4135-y.
Oliveira IG, Moraes AP, Almeida EM, Assis FNM, Cabral JS, Barros F, Felix LP. 2015. Chromosomal evolution in Pleurothallidinae (Orchidaceae: Epidendroideae) with an emphasis on the genus Acianthera: chromosome numbers and heterochromatin. Bot J Linn Soc 178(1): 102–120. https://doi.org/10.1111/boj.12273
Paszko B. 2006. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol 258(1-2): 39-48. https://doi.org/10.1007/s00606-005-0389-2
Pierozzi NI. 2011. Karyotype and NOR-banding of mitotic chromosomes of some Vitis L. species. Rev Bras Frutic 33:564-570. https://doi.org/10.1590/S0100-29452011000500077
Piet Q, Droc G, Marande W, Sarah G, Bocs S, Klopp C, Bourge M, Siljak-Yakovlev S, Bouchez O, Lopez-Roques C, et al. 2022. A chromosome-level, haplotype-phased Vanilla planifolia genome highlights the challenge of partial endoreplication for accurate wholegenome assembly. Plant Commun 3(5): 100330. https://doi.org/10.1016/j.xplc.2022.100330
Romero-Zarco C. 1986. A new method for estimating karyotype asymmetry. Taxon 35(3):526-530. https://doi.org/10.2307/1221906
Silverman BW. 2017. Density estimation for statistics and data analysis. New York (NY): Chapman & Hall.
Souza LGR, Crosa O, Guerra M. 2010. Karyological circumscription of Ipheion Rafinesque (Gilliesioideae, Alliaceae). Plant Syst Evol 287:119-127. https://doi.org/10.1007/s00606-010-0304-3
Souza LGR, Crosa O, Speranza P, Guerra M. 2012. Cytogenetic and molecular evidence suggest multiple origins and geographical parthenogenesis in Nothoscordum gracile (Alliaceae). Ann Bot 109(5): 987-999. https://doi.org/10.1093/aob/mcs020
Stebbins GL. 1971. Chromosomal evolution in higherplants. London: Edward Arnold.
Stedje B. 1989. Chromosome evolution within the Ornithoghlum tenuifolium complex (Hyacinthaceae), with special emphasis on the evolution of bimodal karyotypes. Plant Syst Evol 166: 79-89. https://doi.org/10.1007/BF00937877
Tanaka R. 1967. A comparative karyotype aAnalysis in Haplopappus gracilis (2n=4) and H. ravenii (2n=8). Cytologia 32(3-4): 542-552. https://doi.org/10.1508/cytologia.32.542
Thrun MC, Gehlert T, Ultsch A. 2020. Analyzing the fine structure of distributions. PLoS ONE 15(10): e0238835. https://doi.org/10.1371/journal.pone.0238835
Vaio M, Gardner A, Emshwiller E, Guerra M. 2013. Molecular phylogeny and chromosome evolution among the creeping herbaceous Oxalis species of sections Corniculatae and Ripariae (Oxalidaceae). Mol Phylogenet Evol 68(2):199–211. https://doi.org/10.1016/j.ympev.2013.03.019
Vaio M, Gardner A, Speranza P, Emshwiller E, Guerra M. 2016. Phylogenetic and cytogenetic relationships among species of Oxalis section Articulatae (Oxalidaceae). Plant Syst Evol 302:1253–1265. https://doi.org/10.1007/s00606-016-1330-6
Vanzella ALL, Cuadrado A, Guerra M. 2003. Localization of 45S rDNA and telomeric sites ob holocentric chromosomes of Rhynchospora tenuis Link (Cyperaceae). Genet Mol Biol 26(2): 199-201. https://doi.org/10.1590/S1415-47572003000200014
Violetta K, Pistrick K, Gernand D, Meister A, Ghukasyan A, Gabrielyan I, Houben A. 2005. Characterisation of the low-chromosome number grass Colpodium versicolor (Stev.) Schmalh. (2n = 4) by molecular cytogenetics. Caryologia 58(3): 241-245. https://doi.org/10.1080/00087114.2005.10589457
Vosa C. 1997. Heterochromatin and ecological adaptation in southern African Ornithogalum (Liliaceae). Caryologia 50(2): 97–103. https://doi.org/10.1080/00087114.1997.10797389
Watkins GM. 1936. Chromosome numbers and species characters in Yucca. Am J Bot 23(5): 328–333. https://doi.org/10.2307/2436092
Weiss-Schneeweiss H, Schneeweiss GM. 2013. Karyotype diversity and evolutionary trends in angiosperms. In: Greilhuber J, Dolezel J, Wendel J, editors. Plant genome diversity. Volume 2. Vienna: Springer. p. 209–230.
Wrensch DL, Kethley JB, Norton RA. 1994. Cytogenetics of holokinetic chromosomes and inverted meiosis: keys to the evolutionary success of mites, with generalizations on eukaryotes. In: Houck MA, editor. Mites. Boston: Springer. p. 282–343.
Wu J. 2012. Advances in K-means clustering: a data mining thinking. Springer Theses. Heidelberg: Springer Berlin.
Yang F, O’Brien PCM, Wienberg J, Neitzel H, Lin CC, Ferguson-Smith MA. Chromosomal evolution of the Chinese muntjac (Muntiacus reevesi) Chromosoma 106(1): 37-43. https://doi.org/10.1007/s004120050222
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Leylson Ferreira Araújo, Charlys Seixas Maia Dornelas, Leonardo Pessoa Felix, Felipe Nollet Medeiros de Assis

This work is licensed under a Creative Commons Attribution 4.0 International License.
- Copyright on any open access article in a journal published byCaryologia is retained by the author(s).
- Authors grant Caryologia a license to publish the article and identify itself as the original publisher.
- Authors also grant any third party the right to use the article freely as long as its integrity is maintained and its original authors, citation details and publisher are identified.
- The Creative Commons Attribution License 4.0 formalizes these and other terms and conditions of publishing articles.
- In accordance with our Open Data policy, the Creative Commons CC0 1.0 Public Domain Dedication waiver applies to all published data in Caryologia open access articles.