Karyological variability and chromosomal asymmetry in highland cultivars of Chenopodium quinoa Willd. (Amaranthaceae)
DOI:
https://doi.org/10.13128/caryologia-625Keywords:
Chenopodium quinoa, chromosome, karyotype, chromosomal asymmetry, inter-varietal symmetryAbstract
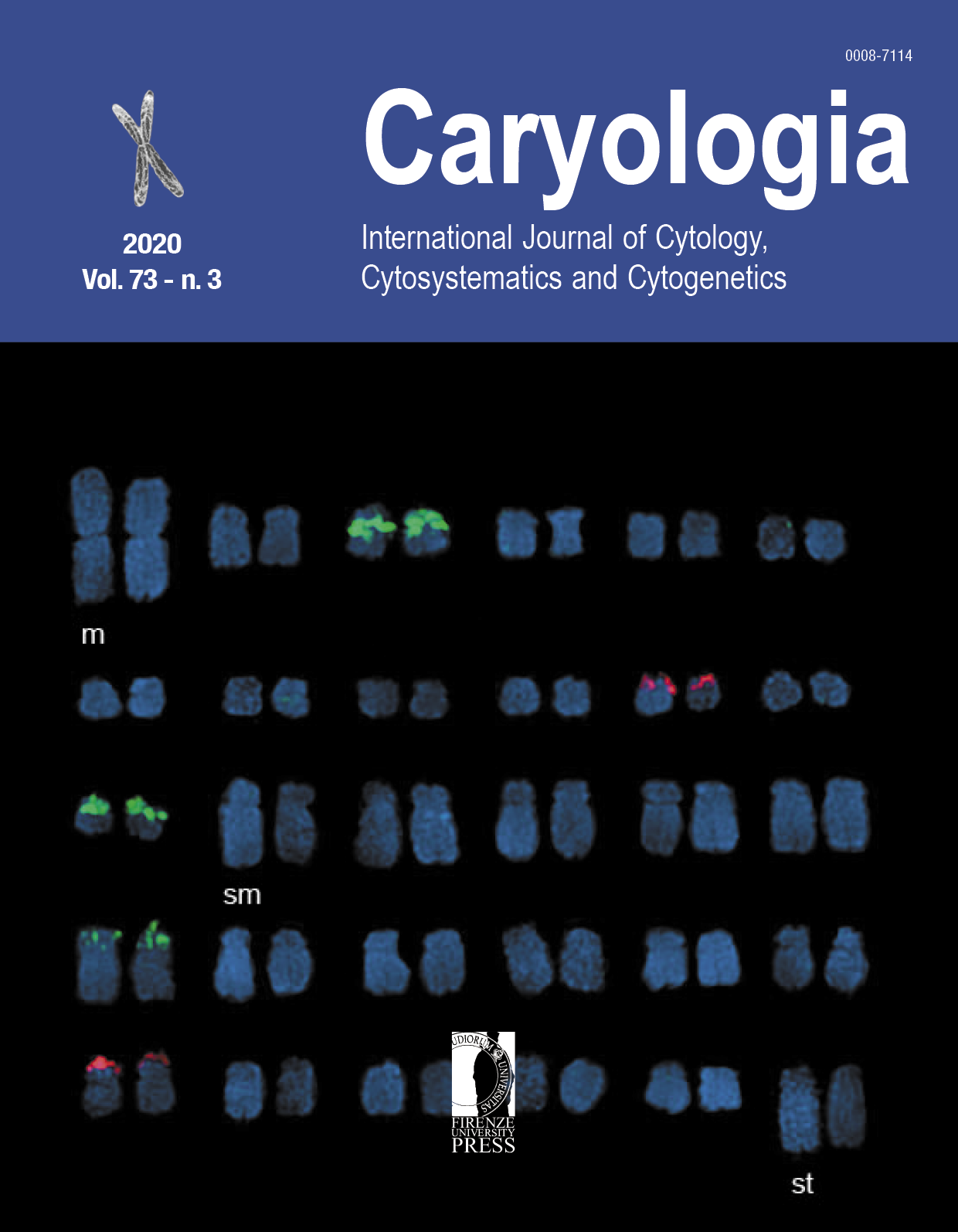
Chenopodium quinoa Willd. is rapidly gaining importance worldwide as a superfood. However, structural diversity and asymmetry analyses of chromosomes of different cultivars of the species are largely understudied primarily owing to their small chromosomes. In this paper, karyomorphological investigations were performed on 21 cultivars of C. quinoa with varying seed morphology cultivated widely in the highland regions of the Andes, which is the center of domestication of this species. Somatic chromosome number was found to be 2n= 36 in all cultivars with no occurrence of mixoploidy. Lengths of individual chromosomes varied between 0.63–6.53 ?m, with their short arms ranging from 0.25–2.95 ?m and long arms between 0.38–3.58 ?m. Types of primary constriction ranged from median to sub–terminal. One pair of chromosome in each complement possessed a secondary constriction. Chromosome complements of all cultivars belonged to the asymmetry class 2B with an average asymmetry index value of 3.21±0.61. Values of intra- and inter-chromosomal asymmetry indices were 0.30±0.02 and 0.20±0.02 respectively across all cultivars. The average coefficients of variation of chromosome lengths was 19.77±2.11 and average centromeric index was 16.28±2.12. Arm ratio of the chromosomes varied from 0.34 to 5.76. The mean values of karyotypic asymmetry, symmetry index and karyotype asymmetry index percentage were 17.69±1.53, 147.69±5.54 and 58.71±0.86 respectively. Pearson correlation revealed strong correlation within inter– and intra–chromosomal asymmetry indices. Our analyses uncovered higher chromosomal variation in quinoa than previously found with high inter–varietal similarities among the studied cultivars, revealed from scattered diagrams between asymmetry indices.
Downloads
References
Arano H. 1963. Cytological studies in subfamily Carduoideae (Compositae) of Japan. IX. The karyotype analysis and phylogenic considerations on Pertya and Ainsliaea. Bot Mag (Tokyo). 76:32-39.
Bennett MD, Leitch IJ. 2005. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot. 95:45-90.
Bhadra S, Bandyopadhyay M. 2015. Karyomorphological investigations on some economically important members of Zingiberaceae from Eastern India. Caryologia. 68:184-192.
Bhargava A, Shukla S, Ohri D. 2006. Karyotypic studies on some cultivated and wild species of Chenopodium (Chenopodiaceae). Genet Resour Crop Evol. 53:1309-1320.
Cárdenas M, Hawkes JG. 1948. Número de cromosomas de algunas plantas nativas cultivadas por los indios en los Andes [Number of chromosomes of some native plants grown by the Indians in the Andes]. Rev de Agricul Bolivia. 4:30-32. Spanish.
Cornille A, Giraud T, Smulders MJM, Roldan-Ruiz I, Gladieux P. 2014. The domestication and evolutionary ecology of apples. Trends Genet. 30:57-65.
Fuentes-Bazan S, Mansion G, Borsch T. 2012. Towards a species level tree of the globally diverse genus Chenopodium (Chenopodiaceae). Mol Phylogenet Evol. 62:359-374.
Gandarillas H. 1979. Botanica. Quinua y kaniwa. Cultivos Andinos [Botany. Quinoa and Kaniwa. Andean Crops]. In: Tapia ME, editor. Serie libros y materiales educativos [Educational books and materials series]. Colombia (Bogotá): Instituto Interamericano de Ciencias Agricolas; p. 20-44. Spanish.
Gandarillas H, Luizaga J. 1967. Número de cromosomas de Chenopodium quinoa Willd. en radículas y raicillas [Number of chromosomes of Chenopodium quinoa Willd. in radicles and rootlets]. Turrialba. 17:275-279. Spanish.
Giusti L. 1970. El género Chenopodium en Argentina: I. Números de cromosomas [The genus Chenopodium in Argentina: I. Numbers of chromosomes]. Darwiniana. 16:98-105. Spanish.
Greilhuber WJ, Speta LF. 1976. C–banded karyotypes in the Scilla hohenackeri group, S. persica, and Puschkinia (Liliaceae). Plant Syst Evol. 126:149-188.
Heiser CB. 1963. Numeración cromosomica de plantas ecuatorianas [Chromosome numbering of Ecuadorian plants]. Ciencia y Nat. (Quito). 6:2-6. Spanish.
Huziwara Y. 1962. Karyotype analysis in some genera of Compositae. VIII. Further studies on the chromosomes of Aster. Am J Bot. 49:116-119.
Jarvis DE, Ho YS, Lightfoot DJ, Schmöckel SM, Li B, Borm TJA, Ohyanagi H, Mineta K, Michell CT, Saber N, et al. 2017. The genome of Chenopodium quinoa. Nature. 542:307-312.
Kawatani T, Ohno T. 1950. Chromosome numbers of genus Chenopodium, I. Chromosome numbers of Mexican Tea (Chenopodium ambrosioides L.), American Wormseed (Chenopodium ambrosioides L. var. anthelminticum A. Gray and some allied species. Jap Jour Genet. 25:177-180.
Kjellmark S. 1934. Einige neue Chromosomenzahlen in der Familie Chenopodiaceae [Some new chromosome numbers in the family Chenopodiaceae]. Bot Notiser. 1934:136-149. German.
Kolano B, Pando LG, Maluszynska J. 2001. Molecular cytogenetic studies in Chenopodium quinoa and Amaranthus caudatus. Acta Soc Bot Pol. 70:85-90.
Kolano B, Tomczak H, Molewska R, Jellen EN, Maluszynska J. 2012. Distribution of 5S and 35S rRNA gene sites in 34 Chenopodium species (Amaranthaceae). Bot J Linn Soc. 170:220-231.
Krak K, Vít P, Belyayev A, Douda J, Hreusová L, Mandák B. 2016. Allopolyploid origin of Chenopodium album s. str. (Chenopodiaceae): A molecular and cytogenetic insight. PLoS ONE. 11:e0161063.
Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52:201-220.
Levitsky GA. 1931. The karyotype in systematics. Bull Appl Bot Gen Pl Breed. 27:220-240.
Mandák B, Krak K, Víta P, Pavlíková Z, Lomonosova MN, Habibi F, Wang L, Jellen EN, Douda J. 2016. How genome size variation is linked with evolution within Chenopodium sensu lato. Perspect Plant Ecol Evol Syst. 23:18-32.
Maughan PJ, Kolano BA, Maluszynska J, Coles ND, Bonifacio A, Rojas J, Coleman CE, Stevens MR, Fairbanks DJ, Parkinson SE, et al. 2006. Molecular and cytological characterization of ribosomal RNA genes in Chenopodium quinoa and Chenopodium berlandieri. Genome. 49:825-839.
Medeiros-Neto E, Nollet F, Moraes AP, Felix LP. 2017. Intrachromosomal karyotype asymmetry in Orchidaceae. Genet Mol Biol. 40:610-619.
Murphy KM, Bazile D, Kellogg J, Rahmanian M. 2016. Development of a worldwide consortium on evolutionary participatory breeding in quinoa. Front Plant Sci. 7:608.
Palomino G, Hernández LT, Torres EDLC. 2008. Nuclear genome size and chromosome analysis in Chenopodium quinoa and C. berlandieri subsp. nuttalliae. Euphytica. 164:221-230.
Pan Y, Wang X, Sun G, Li F, Gong X. 2016. Application of RAD sequencing for evaluating the genetic diversity of domesticated Panax notoginseng (Araliaceae). PLoS ONE. 11:e0166419.
Paszko B. 2006. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol. 258:39-48.
Pereira E, Encina-Zelada C, Barros L, Gonzales-Barron U, Cadavez V, Ferreira ICFR. 2019. Chemical and nutritional characterization of Chenopodium quinoa Willd. (quinoa) grains: a good alternative to nutritious food. Food Chem. 280:110-114.
Peruzzi L, Ero?lu HE. 2013. Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogenet. 7:1-9.
Rahiminejad MR, Gornall RJ. 2004. Flavonoid evidence for allopolyploidy in the Chenopodium album aggregate (Amaranthaceae). Plant Syst Evol. 246:77-87.
Ruiz KB, Biondi S, Oses R, Acuña-Rodríguez IS, Antognoni F, Martinez-Mosqueira EA, Coulibaly A, Canahua-Murillo A, Pinto M, Zurita-Silva A, et al. 2014. Quinoa biodiversity and sustainability for food security under climate change. A review. Agron Sustain Dev. 34:349-359.
Salazar J, Torres ML, Gutierrez B, Torres AF. 2019. Molecular characterization of Ecuadorian quinoa (Chenopodium quinoa Willd.) diversity: implications for conservation and breeding. Euphytica. 215:60.
Stebbins GL. 1971. Chromosomal evolution in higher plants. London (UK): Edward Arnold Publishers Ltd.
Sun W, Ma X, Zhang J, Su F, Zhang Y, Li Z. 2017. Karyotypes of nineteen species of Asteraceae in the Hengduan Mountains and adjacent regions. Plant Diversity. 39:194-201.
Suzuki G, Ogaki Y, Hokimoto N, Xiao L, Kikuchi-Taura A, Harada C, Okayama R, Tsuru A, Onishi M, Saito N, et al. 2012. Random BAC FISH of monocot plants reveals differential distribution of repetitive DNA elements in small and large chromosome species. Plant Cell Rep. 31:621-628.
Tanksley SD, McCouch SR. 1997. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 277:1063-1066.
Wang S, Tsuchiya T, Wilson H. 1993. Chromosome studies in several species of Chenopodium from North and South America. J Genet Breed. 47:163-170.
Wulff HD. 1936. Die polysomatie der Chenopodiaceen. Planta. 26:275-290.
Yangquanwei Z, Neethirajan S, Karunakaran C. 2013. Cytogenetic analysis of quinoa chromosomes using nanoscale imaging and spectroscopy techniques. Nanoscale Res Lett. 8:463.
Zarco CR. 1986. A new method for estimating karyotype asymmetry. Taxon. 35:526-530.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2020 Sreetama Bhadra, Zhi-Quan Cai

This work is licensed under a Creative Commons Attribution 4.0 International License.
- Copyright on any open access article in a journal published byCaryologia is retained by the author(s).
- Authors grant Caryologia a license to publish the article and identify itself as the original publisher.
- Authors also grant any third party the right to use the article freely as long as its integrity is maintained and its original authors, citation details and publisher are identified.
- The Creative Commons Attribution License 4.0 formalizes these and other terms and conditions of publishing articles.
- In accordance with our Open Data policy, the Creative Commons CC0 1.0 Public Domain Dedication waiver applies to all published data in Caryologia open access articles.