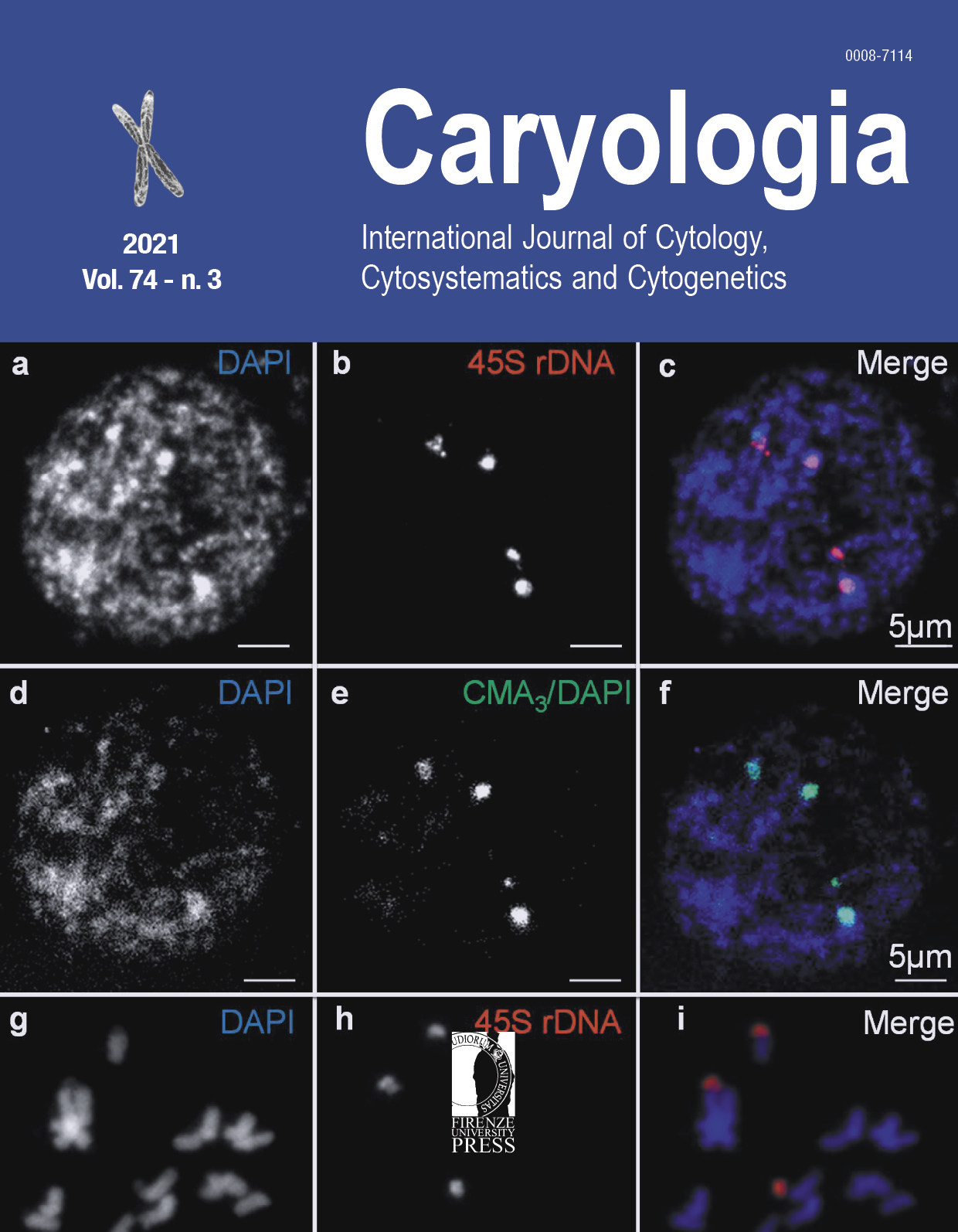
Chromomycin A3 banding and chromosomal mapping of 45S and 5S ribosomal RNA genes in bottle gourd
DOI:
https://doi.org/10.36253/caryologia-1134Keywords:
bottle gourd, chromomycin A3, fluorescence in situ hybridization, local accessions, ribosomal DNA, 45S, 5SAbstract
Ribosomal DNAs and various banding patterns are landmarks in molecular cytogenetics providing useful information for karyotyping and addressing individual chromosomes. Bottle gourd is the only cultivated species of the Lagenaria genus with high genetic diversity. After CMA3/DAPI fluorochrome banding we investigated the GC- and AT-rich regions in interphase nuclei of five different local accessions. Fluorescence in situ hybridization (FISH) was conducted to determine the number and location of 45S and 5S rDNAs in bottle gourd. Our results showed four strong CMA3 regions in interphase and on mitotic metaphase chromosomes. FISH revealed four strong signals of 45S rDNA at the termini of two metaphase chromosome pairs and terminal 5S rDNA signals at another pair of chromosomes. The presence of four positive CMA3 bands colocalizes with four 45S rDNA signals in all bottle gourd accessions. Our results allow distinguishing two out of eleven chromosome pairs of bottle gourd.
Downloads
References
Achigan-Dako EG, Fuchs J, Ahanchede A, Blattner FR (2008) Flow cytometric analysis in Lagenaria siceraria (Cucurbitaceae) indicates correlation of genome size with usage types and growing elevation. Plant Syst Evol 276:9–19. https://doi.org/10.1007/s00606-008-0075-2
Beevy SS, Kuriachan P (1996) Chromosome numbers of south Indian Cucurbitaceae and a note on the cytological evolution in the family. J Cytol Genet 31:65–71
Erickson DL, Smith BD, Clarke AC, et al (2005) An Asian origin for a 10,000-year-old domesticated plant in the Americas. Proc Natl Acad Sci USA 102:18315–18320. https://doi.org/10.1073/pnas.0509279102
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885. https://doi.org/10.1093/nar/7.7.1869
Gerlach WL, Dyer TA (1980) Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res 8:4851–4865. https://doi.org/10.1093/nar/8.21.4851
Han YH, Zhang ZH, Liu JH, et al (2008) Distribution of the tandem repeat sequences and karyotyping in cucumber (Cucumis sativus L.) by fluorescence in situ hybridization. Cytogenet Genome Res 122:80–88. https://doi.org/10.1159/000151320
Hasterok R, Wolny E, Hosiawa M, et al (2006) Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Ann Bot 97:205–216. https://doi.org/10.1093/aob/mcj031
Heslop-Harrison JSP, Schwarzacher T (2011) Organisation of the plant genome in chromosomes. Plant J 66:18–33. https://doi.org/10.1111/j.1365-313X.2011.04544.x
Kim ES, Punina EO, Rodionov AV (2002) Chromosome CPD(PI/DAPI)- and CMA/DAPI-banding patterns in Allium cepa L. Genetika 38:489–496 https://doi.org/10.1023/A:1015250219322
Li K-P, Wu Y-X, Zhao H, et al (2016) Cytogenetic relationships among Citrullus species in comparison with some genera of the tribe Benincaseae (Cucurbitaceae) as inferred from rDNA distribution patterns. BMC Evol Biol 16:85. https://doi.org/10.1186/s12862-016-0656-6
Lombello RA, Pinto-Maglio CAF (2007) Cytomolecular studies in Momordica charantia L. (Cucurbitaceae), a potential medicinal plant. Cytologia 72:415–418. https://doi.org/10.1508/cytologia.72.415
Long EO and Dawid IB (1980) Repeated genes in eukaryotes. Annual Review of Biochemistry 49:727–764. https://doi.org/10.1146/annurev.bi.49.070180.003455
Lysak, M. A., Berr, A., Pecinka, A., Schmidt, R., McBreen, K., & Schubert, I. (2006). Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proceedings of the National Academy of Sciences, 103(13), 5224–5229. https://doi.org/10.1073/pnas.0510791103
Maragheh FP, Janus D, Senderowicz M, et al (2019) Karyotype analysis of eight cultivated Allium species. J Appl Genet 60:1–11. https://doi.org/10.1007/s13353-018-0474-1
Morimoto Y, Mvere B (2004) Lagenaria siceraria. Backhuys Publishers/CTA, Wageningen/Leiden
Probst AV (2018) A compendium of methods to analyze the spatial organization of plant chromatin. Methods Mol Biol 1675:397–418. https://doi.org/10.1007/978-1-4939-7318-7_23
Santos AP, Gaudin V, Mozgová I, et al (2020) Tiding-up the plant nuclear space: domains, function and dynamics. J Exp Bot. https://doi.org/10.1093/jxb/eraa282
Santos-Sanchês, R. de C. V., Souza, M. M., Melo, C. A. F. de, Silva, G. S., Xavier, R. C., Nunes, G. H. de S., & Araújo, I. S. (2019). Karyotypic characterization of melon accessions. Científica, 47(1), 91-103. https://doi.org/10.15361/1984-5529.2019v47n1p91-103
Schweizer D (1976) Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma 58:307–324. https://doi.org/10.1007/BF00292840
Sugiyama K, Kami D, Muro T (2014) Induction of parthenocarpic fruit set in watermelon by pollination with bottle gourd (Lagenaria siceraria (Molina) Standl.) pollen. Scientia Horticulturae 171:1–5. https://doi.org/10.1016/j.scienta.2014.03.008
Tek AL, Kashihara K, Murata M, Nagaki K (2011) Functional centromeres in Astragalus sinicus include a compact centromere-specific histone H3 and a 20-bp tandem repeat. Chromosome Res 19:969–978. https://doi.org/10.1007/s10577-011-9247-y
Waminal NE, Kim HH (2012) Dual-color FISH karyotype and rDNA distribution analyses on four Cucurbitaceae species. Hortic Environ Biotechnol 53:49–56. https://doi.org/10.1007/s13580-012-0105-4
Xu P, Wu X, Luo J, et al (2011) Partial sequencing of the bottle gourd genome reveals markers useful for phylogenetic analysis and breeding. BMC Genomics 12:467. https://doi.org/10.1186/1471-2164-12-467
Volkov RA, Panchuk II, Borisjuk NV, et al (2017) Evolutional dynamics of 45S and 5S ribosomal DNA in ancient allohexaploid Atropa belladonna. BMC Plant Biol 17:21. https://doi.org/10.1186/s12870-017-0978-6
Yamamoto M, Abkenar AA, Matsumoto R, et al (2007) CMA banding patterns of chromosomes in major Citrus species. J Japan Soc Hort Sci 76:36–40. https://doi.org/10.2503/jjshs.76.36
Yeti?ir, H., Kurt, ?., Sari, N., & Tok, F. M. (2007) Rootstock potential of Turkish Lagenaria siceraria germplasm for watermelon: plant growth, graft compatibility, and resistance to Fusarium. Turk J Agric For, 31(6), 381-388. https://doi.org/10.3906/tar-0707-56
Yildiz M, Cuevas HE, Sensoy S, et al (2015) Transferability of Cucurbita SSR markers for genetic diversity assessment of Turkish bottle gourd (Lagenaria siceraria) genetic resources. Biochemical Systematics and Ecology 59:45–53. https://doi.org/10.1016/j.bse.2015.01.006
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2021 Ahmet L. Tek, Hümeyra Y?ld?z, Kamran Khan, Bilge ?. Y?ld?r?m

This work is licensed under a Creative Commons Attribution 4.0 International License.
- Copyright on any open access article in a journal published byCaryologia is retained by the author(s).
- Authors grant Caryologia a license to publish the article and identify itself as the original publisher.
- Authors also grant any third party the right to use the article freely as long as its integrity is maintained and its original authors, citation details and publisher are identified.
- The Creative Commons Attribution License 4.0 formalizes these and other terms and conditions of publishing articles.
- In accordance with our Open Data policy, the Creative Commons CC0 1.0 Public Domain Dedication waiver applies to all published data in Caryologia open access articles.