Divergence in the chromosomal distribution of repetitive sequences in Neotropical cichlid species of the genus Lugubria
DOI:
https://doi.org/10.36253/caryologia-3718Keywords:
repetitive DNAs, Neotropical cichlids, ribosomal genesAbstract
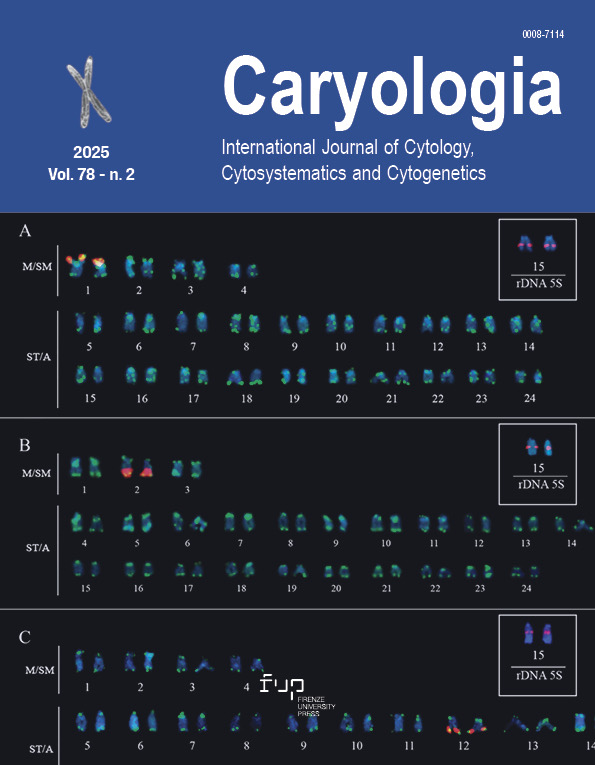
Cytogenetic studies provide valuable insights into the evolutionary dynamics of fish genomes, particularly in groups with high species diversity and ecological relevance. Among Neotropical cichlids, chromosomal data have revealed both conservation patterns and significant structural variations, reflecting intense karyotypic diversification. In this context, mapping repetitive DNA sequences has proven useful in aiding understanding of genomic organization and chromosomal evolution. However, information remains scarce for several cichlid genera. The present study investigated the chromosomal distribution of repetitive sequences, such as 18S and 5S ribosomal genes, as well as telomeric sequences, in three Amazonian species of Lugubria: L. cincta, L. strigata, and L. lugubris. The results revealed a diploid number of 2n = 48, along with variations in the karyotypic formula among the species. Mapping of repetitive sequences revealed distinct patterns of 18S rDNA distribution, with clusters located on different chromosome pairs. Conversely, the 5S rDNA showed a conserved position on a subtelocentric/acrocentric pair in all three species. Furthermore, the presence of interstitial telomeric sequences in L. cincta and L. strigata indicates greater genomic plasticity in these species, suggesting more pronounced chromosome dynamics in the genus Lugubria. These data contribute to the understanding of chromosomal evolution and diversification in this diverse group of Neotropical cichlids and may aid in future cytotaxonomic studies.
Downloads
References
Alanazi AFR, Parkinson GN, Haider S. 2024. Structural motifs at the telomeres and their role in regulatory pathways. Biochemistry. 63(7):827–842. https://doi.org/10.1021/acs.biochem.4c00023.
Arbour JH, López‐Fernández H. 2014. Adaptive landscape and functional diversity of Neotropical cichlids: implications for the ecology and evolution of Cichlinae (Cichlidae; Cichliformes). J Evol Biol. 27(11):2431–2442. https://doi.org/10.1111/jeb.12486.
Ayarpadikannan S, Kim HS. 2014. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 12(3):98–104. https://doi.org/10.5808/GI.2014.12.3.98.
Balshine S, Abate ME. 2021. Parental care in cichlid fishes. In: The behavior, ecology and evolution of cichlid fishes. Cham (CH): Springer. p. 541–586. https://doi.org/10.1007/978-94-024-2080-7_15.
Belyayev A, Kalendar R, Josefiová J, Paštová L, Habibi F, Mahelka V, Mandák B, Krak K. 2023. Telomere sequence variability in genotypes from natural plant populations: unusual block-organized double-monomer terminal telomeric arrays. BMC Genomics. 24(1):96. https://doi.org/10.1186/s12864-023-09657-y.
Bernstein E, Allis CD. 2005. RNA meets chromatin. Genes Dev. 19(14):1635–1655. https://doi.org/10.1101/gad.1324305.
Bolzán AD. 2017. Interstitial telomeric sequences in vertebrate chromosomes: origin, function, instability and evolution. Mutat Res Rev Mutat Res. 773:51–65. https://doi.org/10.1016/j.mrrev.2017.04.002.
Cazaux B, Catalan J, Veyrunes F, Douzery EJP, Britton‐Davidian J. 2011. Are ribosomal DNA clusters rearrangement hotspots? A case study in the genus Mus (Rodentia, Muridae). BMC Evol Biol. 11:124. https://doi.org/10.1186/1471-2148-11-124.
Fricke R, Eschmeyer WN, van der Laan R, editors. 2024. Eschmeyer’s catalog of fishes: genera, species, references. [accessed 2024 Mar 6]. http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp.
Genner MJ. 2023. Cichlid fish seized an ecological opportunity to diversify. Nature. 622(7982):243–244. https://doi.org/10.1038/d41586-023-03014-5.
Gross MC, Schneider CH, Valente GT, Martins C, Feldeberg E. 2010. Variability of 18S rDNA locus among Symphysodon fishes: chromosomal rearrangements. J Fish Biol. 76(5):1117–1127. https://doi.org/10.1111/j.1095-8649.2010.02550.x.
Instituto Chico Mendes de Conservação da Biodiversidade. 2018. Livro vermelho da fauna brasileira ameaçada de extinção: Volume VI – Peixes. Brasília (DF): ICMBio.
Kejnovský E, Jedlička P. 2022. Nucleic acids movement and its relation to genome dynamics of repetitive DNA. BioEssays. 44(4):e2100242. https://doi.org/10.1002/bies.202100242.
Lafuente E, Beldade P. 2019. Genomics of developmental plasticity in animals. Front Genet. 10:720. https://doi.org/10.3389/fgene.2019.00720.
Lee K, Kim D, Kim W. 2021. Regulation of gene expression by telomere position effect. Int J Mol Sci. 22(23):12807. https://doi.org/10.3390/ijms222312807.
Liao X, Zhu W, Zhou J, et al. 2023. Repetitive DNA sequence detection and its role in the human genome. Commun Biol. 6:954. https://doi.org/10.1038/s42003-023-05322-y.
Lower SE, Dion-Côté AM, Clark AG, Barbash DA. 2019. Special issue: repetitive DNA sequences. Genes. 10(11):896. https://doi.org/10.3390/genes10110896.
Lu X, Liu L. 2024. Genome stability from the perspective of telomere length. Trends Genet. 40(2):175–186. https://doi.org/10.1016/j.tig.2023.10.013.
MacGuigan DJ, Krabbenhoft TJ, Harrington R, Wainwright DK, Backenstose NJC, Near TJ. 2023. Lacustrine speciation associated with chromosomal inversion in a lineage of riverine fishes. Evolution. 77(7):1505–1521. https://doi.org/10.1093/evolut/qpad067.
Majtánová Z, Indermaur A, Nyom ARB, Ráb P, Musilova Z. 2019. Adaptive radiation from a chromosomal perspective: evidence of chromosome set stability in cichlid fishes (Cichlidae: Teleostei) from the Barombi Mbo Lake, Cameroon. Int J Mol Sci. 20(20):4994. https://doi.org/10.3390/ijms20204994.
Marajó L, Viana PF, Ferreira AMV, Py‐Daniel LHR, Cioffi MdB, Sember A, Feldberg E. 2022. Chromosomal rearrangements and the first indication of an ♀X1X1X2X2/♂X1X2Y sex chromosome system in Rineloricaria fishes (Teleostei: Siluriformes). J Fish Biol. 102(2):443–454. https://doi.org/10.1111/jfb.15275.
Martins C, Vicari MR. 2012. Hibridização in situ em cromossomos de peixes. In: Guerra M, editor. Citogenética molecular: protocolos comentados. Ribeirão Preto (SP): Sociedade Brasileira de Genética. p. 89–106.
Matschiner M, Böhne A, Ronco F, Salzburger W. 2020. The genomic timeline of cichlid fish diversification across continents. Nat Commun. 11:5895. https://doi.org/10.1038/s41467-020-17827-9.
Molina WF, Galetti PM Jr. 2002. Robertsonian rearrangements in the reef fish Chromis (Perciformes, Pomacentridae) involving chromosomes bearing 5S rRNA genes. Genet Mol Biol. 25(4):373–377. https://doi.org/10.1590/S1415-47572002000400004.
Ocalewicz K. 2013. Telomeres in fishes. Cytogenet Genome Res. 141(2–3):114–125. https://doi.org/10.1159/000354278.
Paiz LM, Gavazzoni M, Antoniazi GJ, Baumgärtner L, Graça WJ, Feldberg E, Lui RL, Margarido VP. 2024. Trends in chromosome evolution in Crenicichlina (Cichliformes, Cichlidae, Cichlinae): a new perspective based on the recent classification of the pike cichlids. Rev Fish Biol Fish. 34(2):849–866. https://doi.org/10.1007/s11160-024-09842-6.
Šatović-Vukšić E, Plohl M. 2023. Satellite DNAs—From localized to highly dispersed genome components. Genes. 14(3):742. https://doi.org/10.3390/genes14030742.
Singh P, Irisarri I, Torres‐Dowdall J, Thallinger G, Svardal H, Lemmon EM, Lemmon AR, Koblmüller S, Meyer A, Sturmbauer C. 2022. Phylogenomics of trophically diverse cichlids disentangles processes driving adaptive radiation and repeated trophic transitions. Ecol Evol. 12(7):e9077. https://doi.org/10.1002/ece3.9077.
Suarez P, Barroso ICGP, Silva DS, Milhomem SSR, Cabral-De-Mello DC, Martins C, Pieczarka JC, Nagamachi CY. 2017. Highest diploid number among Gymnotiformes: first cytogenetic insights into Rhabdolichops (Sternopygidae). Zebrafish. 14(3):272–279. https://doi.org/10.1089/zeb.2016.1405.
Torres‐Dowdall J, Karagic N, Härer A, Meyer A. 2021. Diversity in visual sensitivity across Neotropical cichlid fishes via differential expression and intraretinal variation of opsin genes. Mol Ecol. 30(8):1880–1891. https://doi.org/10.1111/mec.15855.
Turner GF. 2007. Adaptive radiation of cichlid fish. Curr Biol. 17(19):R827–R831. https://doi.org/10.1016/j.cub.2007.07.026.
Varella HR, Kullander S, Menezes NA, Oliveira C, López‐Fernández H. 2023. Revision of the generic classification of pike cichlids using an integrative phylogenetic approach (Cichlidae: tribe Geophagini: subtribe Crenicichlina). Zool J Linn Soc. 198(4):982–1034. https://doi.org/10.1093/zoolinnean/zlad021.
Vicari MR, Bruschi DP, Cabral-de-Mello DC, Nogaroto V. 2022. Telomere organization and the interstitial telomeric sites involvement in insects and vertebrates chromosome evolution. Genet Mol Biol. 45(suppl 3):e20220071. https://doi.org/10.1590/1678-4685-gmb-2022-0071.
Wang W, Zhang X, Garcia S, Leitch AR, Kovařı́k A. 2023. Intragenomic rDNA variation – the product of concerted evolution, mutation, or something in between? Heredity. 131(3):179–188. https://doi.org/10.1038/s41437-023-00634-5.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Frade, LFS, Santos, CEV, Almeida, BRR, Nagamachi, CY, Pieczarka, JC, Nascimento, LAS, Martins, C, Cardoso, AL, Noronha, RCR

This work is licensed under a Creative Commons Attribution 4.0 International License.
- Copyright on any open access article in a journal published byCaryologia is retained by the author(s).
- Authors grant Caryologia a license to publish the article and identify itself as the original publisher.
- Authors also grant any third party the right to use the article freely as long as its integrity is maintained and its original authors, citation details and publisher are identified.
- The Creative Commons Attribution License 4.0 formalizes these and other terms and conditions of publishing articles.
- In accordance with our Open Data policy, the Creative Commons CC0 1.0 Public Domain Dedication waiver applies to all published data in Caryologia open access articles.