Cytogenetics of Diphysa americana (Mill.) M.Sousa (Leguminosae-Papilionoideae-dalbergioid clade), a rare species from the coast of Oaxaca, Mexico
DOI:
https://doi.org/10.36253/caryologia-2300Keywords:
basic number, dalbergioid clade, karyotype, SAT-chromosomesAbstract
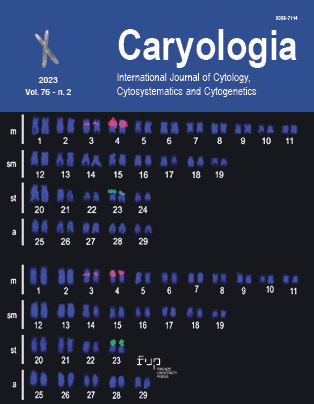
Diphysa Jacq. is an essentially Mexican and Central American genus that includes 21 species and only one cytogenetic report. In this work, a surface spread and air-drying method was used to obtain the karyotype of Diphysa americana (Mill) M.Sousa, a rare native tree that grows in a coastal town in the State of Oaxaca, Mexico. Metaphase cells showed a 2n = 20, consistent with the predominant diploid number in the dalbergioid clade. This number contrasts with a previously reported 2n = 16. The karyotypic formula 5m + 5sm, first proposed for a species of the genus, denotes a slightly asymmetric karyotype. The presence of secondary constrictions associated with satellites on the short arms of a pair of sm chromosomes and other cytogenetic parameters require studies in other species of the genus to verify their taxonomic utility. In addition, cells in prometaphase exhibited a circular fragment of unknown origin like that observed in a species of the genus Aeschynomene, also dalbergioid. This fragment could be related to extrachromosomal circular DNA (eccDNA) observed in other plants. Diphysa is a small, cytogenetically favorable genus, and further studies will exhibit the karyotypic diversity that underlies its diversification.
Downloads
References
Acosta I. 1993. Lluvia de semillas en matorrales de dunas costeras en el Morro de La Mancha Veracruz. Bachelor of Science Thesis. Facultad de Ciencias. Universidad Nacional Autónoma de México, México, DF.
Atchison E. 1951. Studies in the Leguminosae. VI. Chromosome numbers among tropical woody species. Am J Bot. 38:538-546. https://doi.org/10.1002/j.1537-2197.1951.tb14855.x
Biondo E, Miotto STS, Schifino-Wittmann MT. 2006. Cytogenetics of species of Chamaecrista (Leguminosae-Caesalpinioideae) native to southern Brazil. Bot J Linn Soc. 150(4):429-439. https://doi.org/10.1111/j.1095-8339.2006.00480.x
Cardoso D, de Queiroz LP, Pennington RT, de Lima HC, Fonty E, Wojciechowski MF, Lavin M. 2012. Revisiting the phylogeny of papilionoid legumes: new insights from comprehensively sampled early-branching lineages. Am J Bot. 99:1991-2013. http://www.jstor.org/stable/23321299.
Cardoso D, Pennington RT, de Queiroz LP, Boatwright JS, Van Wyk B-E, Wojciechowski MF, Lavin M. 2013. Reconstructing the deep-branching relationships of the papilionoid legumes. S Afr J Bot. 89:58-75. https://doi.org/10.1016/j.sajb.2013.05.001
Cardoso D, Mattos CMJ, Filardi F, Delgado-Salinas A, Lavin M, de Moraes PLR, Tapia-Pastrana F, de Lima HC. 2020. A molecular phylogeny of the pantropical papilionoid legume Aeschynomene supports reinstating the ecologically and morphologically coherent genus Ctenodon. Neodiversity, 13:1-38. https://doi.org/10.13102/neod/131.1
Cervantes A, Linares J, Quintero E. 2019. An updated checklist of the Mexican species of Dalbergia (Leguminosae) to aid in its conservation efforts. Rev Mex Biodivers. 90: e902528. https://doi.org/10.22201/ib.20078706e.2019.90.2528
CONABIO (Comisión Nacional para el Conocimiento y uso de la Biodiversidad). [accessed 2023 March 23]. https://enciclovida.mx/especies/185459
Cohen S, Houben A, Segal D. 2008. Extrachromosomal circular DNA derived from tandemly repeated genomic sequences in plants. Plant J. 53:1027-1034. https://doi.org/10.1111/j.1365-313X.2007.03394.x
Cordeiro JMP, Kaehler M, Souza LG, Felix LP. 2020. Heterochromatin and numeric chromosome evolution in Bignoniaceae, with emphasis on the Neotropical clade Tabebuia alliance. Genet Mol Biol. 43: e20180171. https://doi.org/10.1590%2F1678-4685-GMB-2018-0171
Doyle JJ, Luckow MA. 2003. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 131:900-911. https://doi.org/10.1104%2Fpp.102.018150
Goldblatt P. 1981. Cytology and the phylogeny of Leguminosae. In: Polhill RM, Raven PH, editors. Advances in legume systematics. part 2. Royal Botanic Garden, London: Kew Publishing; p. 427-463.
Huziwara Y. 1962. Karyotype analysis in some genera of Compositae. VIII. Further studies on the chromosomes of Aster. Am J Bot. 49:116-119. https://doi.org/10.2307/2439026
IUCN (Red List of Threatened Species). Version 3.1. [accessed 2023 March 27]. https://www.iucnredlist.org/species/144264286/149030506
Jena S, Sahoo P, Mohanty S, Das AB. 2004. Identification of RAPD markers, in situ DNA content and structural chromosomal diversity in some legumes of the mangrove flora of Orissa. Genetica. 122:217-226. http://dx.doi.org/10.1007/s10709-004-2040-5
Lavin M, Pennington RT, Klitgaard BB, Sprent JI, de Lima HC, Gasson PE. 2001. The Dalbergioid legumes (Fabaceae): Delimitation of a pantropical monophyletic clade. Am J Bot. 88:503-533. https://doi.org/10.2307/2657116
Lavin M, Herendeen PS, Wojciechowski MF. 2005. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst Biol. 54:575-594. https://doi.org/10.1080/10635150590947131
Lavin M, Thulin M, Labat J-N, Pennington RT. 2000. Africa, the odd man out: molecular biogeography of dalbergioid legumes (Fabaceae) suggests otherwise. Syst Bot. 25(3):449-467. https://doi.org/10.2307/2666689
Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas 52:201-219. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x
Lewis G, Schrire B, Mackinder B, Lock M, editors. 2005. Legumes of the world. Richmond, U. K.: Royal Botanic Gardens, Kew.
Lewis GP, Wood JRI, Lavin M. 2012. Steinbachiella (Leguminosae: Papilionoideae: Dalbergieae), endemic to Bolivia, is reinstated as an accepted genus. Kew Bull. 67:789-796. https://doi.org/10.1007/s12225-012-9415-z
Lewke Bandara N, Papini A, Mosti S, Brown T, Smith LMJ. 2013. A phylogenetic analysis of genus Onobrychis and its relationships within the tribe Hedysareae (Fabaceae). Turk J Bot. 37:981-992. https://doi.org/10.3906/bot-1210-32
Lim KB, Wennekes J, de Jong JH, Jacobsen E, van Tuyl JM. 2001. Karyotype analysis of Lilium longiflorum and Lilium rubellum by chromosome banding and fluorescence in situ hybridisation. Genome, 44(5):911-918. https://doi.org/10.1139/g01-066
Lima de Faria A. 1976. The chromosome field. I. Prediction of the location of ribosomal cistrons. Hereditas, 83(1):1-22. https://doi.org/10.1111/j.1601-5223.1976.tb01565 .
LPWG (Legume PhylogenyWorking Group 2013). Legume phylogeny and classification in the 21st century: progress, prospects, and lessons for other species-rich clades. Taxon, 62:217-248. http://dx.doi.org/10.12705/622.8
Manzanero-Medina GI, Vásquez-Dávila MA, Lustre-Sánchez H, Pérez-Herrera A. 2020. Ethnobotany of food plants (quelites) sold in two traditional markets of Oaxaca, Mexico. S Afr J Bot. 130:215-233. https://doi.org/10.1016/j.sajb.2020.01.002
Martín G, Milera M, Iglesias J, Simón L, Hernández H. 2000. Sistemas silvopastoriles para la producción ganadera en Cuba. Intensificación de la ganadería en Centroamérica, beneficios económicos y ambientales. CATIE-FAO. Costa Rica 247 p.
Martins MB, Agostinetto D, Fogliatto S, Vidotto F, Andres A. 2021. Aeschynomene spp. Identification and weed management in rice fields in Southern Brazil. Agronomy, 11:453. https://doi.org/10.3390/agronomy11030453
McMahon MM, Sanderson MJ. 2006. Phylogenetic supermatrix analysis of GenBank sequences from 2228 papilionoid legumes. Syst. Biol. 55:818-836. https://doi.org/10.1080/10635150600999150
Moraes AP, Vatanparast M, Polido C, Marques A, Souza G, Fortuna‑Perez AP, Forni‑Martins ER. 2020. Chromosome number evolution in dalbergioid legumes (Papilionoideae, Leguminosae). Braz J Bot. 43:575-587. https://doi.org/10.1007/s40415-020-00631-6
Nakamura R, Kitamura S, Inoue M, Ohmido N, Fukui K. 2001. Karyotype analysis of Nicotiana kawakamii Y. Ohashi using DAPI banding and rDNA FISH. Theor Appl Genet. 102(6-7):810-814. https://doi.org/10.1007/s001220100577
Pascual-Mendoza S, Manzanero-Medina GI, Saynes-Vásquez A, Vásquez-Dávila MA. 2020. Agroforestry systems of a Zapotec community in the Northern Sierra of Oaxaca, Mexico. Bot Sci. 98(1):128-144. https://doi.org/10.17129/botsci.2423
Pennington RT, Lavin M, Ireland H, Klitgaard B, Preston J, Hu JM. 2001. Phylogenetic relationships of basal papilionoid legumes based upon sequences of the chloroplast trnL intron. Syst. Bot. 26:537-556. http://dx.doi.org/10.1043/0363-6445-26.3.537
Ramírez-Pinero M. 2012. Técnicas para la restauración de la selva baja caducifolia en el centro de Veracruz [master’s thesis]. Xalapa, Veracruz: Instituto de Ecología, A.C. México.
Ramírez-Pinero M, Lira-Noriega A, Guevara S. 2018. Canopy asymmetry in solitary Diphysa americana trees: wind and landscape on the Mexican coast. J Coast Conserv. 23:163-172. https://doi.org/10.1007/s11852-018-0648-3
Rojas-Rodríguez F, Torres-Córdoba G. 2018. Árboles del Valle Central de Costa Rica: reproducción guachipelín (Diphysa americana (Mill.) M. Sousa). RFMK. 16(38): 69-71. https://doi.org/10.18845/rfmk.v16i38.3998
Rzedowski J, Calderón de Rzedowski G, Torres Colín L, Grether R. 2016. Familia Leguminosae. Subfamilia Papilionoideae (Aeschynomene - Diphysa). In: Rzedowski J, Calderón de Rzedowski G, editors. Flora del Bajío y regiones adyacentes. Fascículo 192. Instituto de Ecología A.C., Michoacán, México; p.1-326.
Sousa MS. 1990. Adiciones a las papilionadas de la Flora de Nicaragua y una nueva combinación para Oaxaca, México. Ann Mo Bot Gard. 77: 573-577. https://doi.org/10.2307/2399521
Tapia-Pastrana F. 2012. Karyological characterisation of four American species of Crotalaria (Leguminosae: Papilionoideae) by splash method. Kew Bull. 67(3):427-433. https://doi.org/10.1007/s12225-012-9385-1
Tapia-Pastrana F, Delgado-Salinas A, Caballero J. 2020. Patterns of chromosomal variation in Mexican species of Aeschynomene (Fabaceae, Papilionoideae) and their evolutionary and taxonomic implications. Comp Cytogenet. 14(1):157-182. https://doi.org/10.3897%2FCompCytogen.v14i1.47264
Tapia-Pastrana F, Mercado-Ruaro P. 2001. A combination of the “squash” and “splash” techniques to obtain the karyotype and asses meiotic behavior of Prosopis laevigata L. (Fabaceae: Mimosoideae). Cytologia, 66:11-17. http://dx.doi.org/10.1508/cytologia.66.11
Tapia-Pastrana F, Tapia-Aguirre F. 2018. Localización de satélites y cromosomas NOR para la interpretación del cariotipo de Sesbania virgata (Papilionoideae, Sesbanieae) de dos poblaciones americanas. Bot Sci. 96(4):619-627. https://doi.org/10.17129/botsci.1972
Villaseñor JL. 2016. Checklist of the native vascular plants of Mexico. Rev Mex Biodivers. 87(3):559-902. https://doi.org/10.1016/j.rmb.2016.06.017
WFO (The World Flora Online). [accessed 2023 March 23]. http://www.worldfloraonline.org/taxon/wfo-0000210419
Wojciechowski MF, Lavin M, Sanderson MJ. 2004. A phylogeny of legumes (Leguminosae) based on analysis of the plastid MatK gene resolves many well-supported subclades within the family. Am J Bot. 91:1846-1862. https://doi.org/10.3732/ajb.91.11.1846
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Fernando Tapia-Pastrana

This work is licensed under a Creative Commons Attribution 4.0 International License.
- Copyright on any open access article in a journal published byCaryologia is retained by the author(s).
- Authors grant Caryologia a license to publish the article and identify itself as the original publisher.
- Authors also grant any third party the right to use the article freely as long as its integrity is maintained and its original authors, citation details and publisher are identified.
- The Creative Commons Attribution License 4.0 formalizes these and other terms and conditions of publishing articles.
- In accordance with our Open Data policy, the Creative Commons CC0 1.0 Public Domain Dedication waiver applies to all published data in Caryologia open access articles.