The genome of the southern short-horned tree dragon Acanthosaura meridiona Trivalairat, Sumontha, Kunya & Chaingkul, 2022 (Squamata, Draconinae) was analyzed using classical and molecular techniques to identify and study its chromosomal and repetitive elements
DOI:
https://doi.org/10.36253/caryologia-2961Keywords:
Acanthosaura meridional, Chromosome marker, fluorescence in situ hybridization (FISH), Microsatellite pattern, DraconinaeAbstract
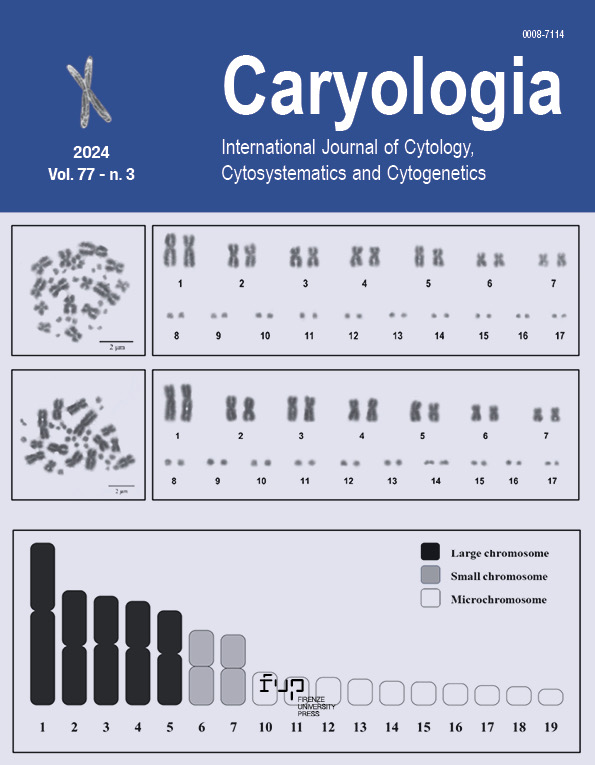
The cytogenetics of the southern short-horned tree dragon (Acanthosaura meridiona) are not reported yet. This study describes the karyotype of Acanthosaura meridiona Trivalairat, Sumontha, Kunya & Chaingkul, 2022 from southern Thailand. We using Giemsa staining, Ag-NOR banding, and fluorescence in situ hybridization (FISH) techniques using microsatellites d(CA)15, d(TA)15, d(CGG)10, and d(CAA)10 probes to analyze the chromosome. The karyotype of the A. meridiona is 2n = 34 chromosomes (fundamental number of 46), of which 5 pairs were large metacentric chromosomes, 2 pairs small metacentric chromosomes, and 20 microchromosomes (chromosome formula: 2n=34=Lm10+ Sm4+20mi). There are no sex differences in karyotypes between males and females. The NORs loci were on pair 5 of the large metacentric macrochromosomes. The FISH technique showed d(CA)15 and d(CGG)10 repeats on specific regions microchromosomes, while signals of d(TA)15 and d(CAA)10 repeats interspersed on macro- and microchromosomes. This study is significant for enhances our comprehension of the evolutionary mechanism of agamid lizards and promotes the conservation of biodiversity in tropical rainforests.
Downloads
References
Adegoke JA, Ejere VC. 1991. Description of the chromosomes of three lizard species belonging to the genus Mabuya (Scincidae, Reptilia). Caryologia. 44:333–342.
Aiumsumang S. Phimphan S. Suwannapoom C, Chaiyasan P, Supiwong W, Tanomtong A. 2021. A comparative chromosome study on five Minnow fishes (Cyprinidae, Cypriniformes) in Thailand. Caryologia 74(1):89-96.
Alam SM, Sarre SD, Georges A, Ezaz T. 2021. Karyotype Characterisation of two Australian dragon lizards (Squamata: Agamidae: Amphibolurinae) reveals subtle chromosomal rearrangements between related species with similar karyotypes. Cytogenet Genome Res. 160(10):610-624.
Alam SM, Sarre SD, Gleeson D, Georges A, Ezaz T. 2018. Did lizards follow unique pathways in sex chromosome evolution? Genes. 9(239).
Alföldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, and et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 477:587–591.
Ananjeva NB, Ermakov OA, Nguyen SN, Nguyen TT, Murphy RW, Lukonina SA, Orlov NL. 2020. A new species of Acanthosaura Gray, 1831 (Squamata: Agamidae) from central highlands, Vietnam. Russ J Herpetol. 27(4):217–230.
Ananjeva NB, Orlov NL, Kalyabina-Hauf SA. 2008. Species of Acanthosaura Gray, 1831 (Agamidae: Sauria, Reptilia) of Vietnam: results of Molecular and Morphological study. Biol Bull. 35(2):178 –186.
Badenhorst D, Hillier LW, Literman R, Montiel EE, Radhakrishnan S, Shen Y, Minx P, Janes DE, Warren WC, Edwards SV, and et al. 2015. Physical mapping and refinement of the painted turtle genome (Chrysemys picta) inform amniote genome evolution and challenge turtle-bird chromosomal conservation. Genome Biol Evol. 7:2038–2050.
Chistiakov DA, Hellemans B, Volckaert FAM. 2006. Microsatellites and their genomic distribution, evolution, function and applications: a review with special reference to fish genetics. Aquaculture. 255:1–29. doi: 10.1016/j.aquaculture.2005.11.031.
Chan-Ard T, Nabhitabhata J, Parr JW. 2015. A field guide to the reptiles of Thailand. NY: Oxford University Press.
Das I. 2015. Field guide to the reptiles of South-East Asia. London: Bloomsbury Publishing.
Deakin JE, Edwards MJ, Patel H, O’Meally D, Lian J, Stenhouse R, Ryan S, Livernois AM, Azad B, Holleley CE, and et al. 2016. Anchoring genome sequence to chromosomes of the central bearded dragon (Pogona vitticeps) enables reconstruction of ancestral squamate macrochromosomes and identifies sequence content of the Z chromosome. BMC Genomics. 17(447).
Deakin JE, Ezaz T. 2019. Understanding the evolution of reptile chromosomes through applications of combined cytogenetics and genomics approaches. Cytogenet. Genome Res. 157:7–20. doi: 10.1159/000495974.
Diong CH, Low MH, Tan EC, Yong HS, Hikida T, Ota H. 2000. On the Monophyly of the Agamid Genus Gonocephalus Kaup, 1825 (Reptilia: Squamata) A Chromosomal Perspective. Curr. Herpetol. 19(2):71-79.
Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 5:435–445. doi: 10.1038/nrg1348.
Grismer LL. 2011. Lizards of Peninsular Malaysia, Singapore and their Adjacent Archipelagos, Edition Chimaira, Frankfürt am Main.
Howell WM, Black DA. 1980. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia. 36: 1014-1015.
Khawporntip W, Jantarat S, Supanuam P, Buatip S, Kraiprom T, Donbundit N, Thongnetr W, Tanomtong A. 2024. First Molecular Cytogenetics Report of Blue-headed Flying Lizard (Draco volans) outside Protected Area of Hala-bala Forest at Than To District, Yala Province, Thailand. Burapha Science Journal, 29(2), 782–792.
Kritpetcharat O, Kritpetcharat C, Luangpirom A, Watcharanon P. 1999. Karyotype of four Agamidae species from the Phu Phan national park in Thailand. Sci. Asia, 25(4):185-188.
Kubat Z, Hobza R, Vyskot B, Kejnovsky E. 2008. Microsatellite accumulation in the Y chromosome of Silene latifolia. Genome 51:350–356.
Li Shu-shen, Wang Ying-xiang, Wang Rui-fang, Li Chong-yun, Liu Guang-zuo. 1981. A Karyotypical Study of Japalura varcoae (Boulenger). Zoological Research, 2(3): 223-228.
Liu S, Hou M, Mo MZ, Rao DQ. 2020. A new species of the genus Acanthosaura (Squamata, Agamidae) from Yunnan, China, with comments on its conservation status. ZooKeys. 959:113–135.
Liu S, Rao D, Hou M, Orlov NL, Ananjeva NB, Zhang D. 2022. Two new species of Acanthosaura Gray, 1831 (Reptilia: Agamidae) from Yunnan province, China Russ J Herpetol. 29(2): 93–109. doi: 10.30906/1026-2296-2022-29-2-93-109.
Manthey U. 2008. Agamid Lizards of Southern Asia-Agamen des südlichen Asien-Draconinae 1. Terralog. Vol. 7a, Edition Chimaira, Frankfürt am Main.
Mezzasalma M, Guarino FM, Odierna G. 2021. Lizards as Model Organisms of Sex Chromosome Evolution: What We Really Know from a Systematic Distribution of Available Data? Genes. 12(1341).
Mezzasalma M, Macirella R, Odierna G, Brunelli E. 2024. Karyotype Diversification and Chromosome Rearrangements in Squamate Reptiles. Genes. 15(371). doi: org/10.3390/genes15030371.
O’Meally D, Miller H, Patel HR, Graves JA, Ezaz T. 2009. The first cytogenetic map of the tuatara, Sphenodon punctatus. Cytogenet Genome Res. 127:213–23.
Ota, H. 1988. Karyotypic differentiation in an agamid lizard, Japalura swinhonis swinhonis. Experientia, 44(1):66-68.
Ota H, Hikida T. 1989. Karyotypes of three species of the genus Draco (Agamidae: Lacertilia) from Sabah, Malaysia. Japan J. Herpetol. 13(1):1-6.
Ota H, Diong CH, Tan EC, Yong HS. 2002. Karyotypes of four agamid lizards from Southeast Asia. Curr. Herpetol. 21(1):35-41.
Patawang I, Pinthong K, Thongnetr W, Sornnok S, Kaewmad P, Tanomtong A. 2018. Additional description of karyotype and meiotic features of Takydromus sexlineatus (Squamata, Lacertidae) from Northeastern Thailand. The Nucleus 61(2):163-169.
Patawang I, Prasopsin S, Suwannapoom C, Tanomtong A, Keawmad P, Thongnetr W. 2022. Chromosomal description of three Dixonius (Squamata, Gekkonidae) from Thailand. Caryologia 75(2):101-108.
Patawang I, Tanomtong A, Chuaynkern Y, Chuaynkern C, Duengkae P. 2015. Karyotype homology between Calotes versicolor and C. mystaceus (Squamata, Agamidae) from northeastern Thailand. The Nucleus 58(2):117-123.
Patawang I, Tanomtong A, Getlekha N, Phimphan S, Pinthong K, Neeratanaphan L. 2017. Standardized karyotype and idiogram of Bengal monitor lizard, Varanus bengalensis (Squamata, Varanidae). Cytologia 82(1):75-82.
Patawang I, Tanomtong A, Kaewmad P, Chuaynkern Y, Duengkae P. 2016. New record on karyological analysis and first study of NOR localization of parthenogenetic brahminy blind snake, Ramphotyphlops braminus (Squamata, Typhlopidae) in Thailand. The Nucleus 59(1): 61-66.
Prasopsin S., Muanglen,N, Ditcharoen S, Suwannapoom C, Tanomtong A, Thongnetr W. 2022. First Report on Classical and Molecular Cytogenetics of Doi Inthanon Benttoed Gecko, Cyrtodactylus inthanon Kunya et al., 2015 Squamata: Gekkonidae) in Thailand. Caryologia 75(2):109-117.
Sassi FMC, Toma GA, Cioffi MB. 2023. “FISH—In fish chromosomes,” in Cytogenetics and molecular cytogenetics. Editor T. Liehr (Boca Raton, FL: CRC Press), 281–296.
Sharma GP, Nakhasi U. 1980. Karyotypic homology and evolution of the Agamid lizards. Cytologia, 45(1-2):
-219.
Silva M, Pereira HS, Bento M, Santos AP, Shaw P, Delgado M, Neves N, Viegas W. 2008. Interplay of ribosomal DNA loci in nucleolar dominance: dominant NORs are upregulated by chromatin dynamics in the wheatrye system. PLoS ONE. 3. doi: 10.1371/journal.pone.0003824.
Singh AK, Banerjee R. 2004. Chromosomal diversity of Indian mammals, amphibians and reptiles. Rec. zool. Surv. India 102(3-4):127-138.
Solleder E, Schmid M. 1988. Cytogenetic studies on Sauria (Reptilia). I. Mitotic chromosomes of the Agamidae. Amphibia-reptilia 9(3):301-310.
Srikulnath K, Matsubara K, Uno Y, Thongpan A, Suputtitada S, Apisitwanich S, Matsuda Y, Nishida C. 2009. Karyological characterization of the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Squamata) by molecular cytogenetic approach. Cytogenet Genome Res, 125(3):213-223.
Srikulnath K, Uno Y, Nishida C, Ota H, Matsuda Y. 2015. Karyotype reorganization in the Hokou gecko (Gekko hokouensis, Gekkonidae): the process of microchromosome disappearance in Gekkota. PLoS One. doi: 10.1371/journal.pone.0134829.
Tautz D, Renz M.1984. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 25:4127–4138. doi: 10.1093/nar/12.10.4127.
Thongnetr W, Aiumsumang S, Tanomtong A, Phimphan S. 2022a. Classical chromosome features and microsatellites repeat in Gekko petricolus (Reptilia, Gekkonidae) from Thailand. Caryologia 75(2):81-88.
Thongnetr W, Prasopsin S, Aiumsumang S, Ditcharoen S, Tanomtong A, Wongchantra P, Bunnaen W, Phimphan S. 2022b. First report of chromosome and karyological analysis of Gekko nutaphandi (Gekkonidae, Squamata) from Thailand: Neo-diploid chromosome number in genus Gekko. Caryologia 75(4): 103-109. doi: 10.36253/caryologia-1875.
Thongnetr W, Tanomtong A, Prasopsin S, Maneechot N, Pinthong K, Patawang I. 2019. Cytogenetic study of the Bent-toed Gecko (Reptilia, Gekkonidae) in Thailand; I: Chromosomal classical features and NORs characterization of Cyrtodactylus kunyai and C. interdigitalis. Caryologia 72(1): 23-28. doi: 10.13128/cayologia-248.
Trifonov VA, Paoletti A, Barucchi VC, Kalinina T, O’Brien PCM, Ferguson-Smith MA Giovannotti M. 2015. Comparative Chromosome Painting and NOR Distribution Suggest a Complex Hybrid Origin of Triploid Lepidodactylus lugubris (Gekkonidae). PLoS ONE. 10. doi: doi.org/10.1371/journal.pone.0132380.
Trivalairat P, Sumontha M, Kunya K, Chiangkul K. 2022. Acanthosaura meridiona sp. nov. (Squamata: Agamidae), a new short-horned lizard from southern Thailand. Herpetological Journal. 32, 34-50. doi: 10.33256/32.1.3450.
Turpin R, Lejeune J. 1965. Les chromosomes humains (caryotype normal et variations pathologiques). Paris: Gauthier-Villars. [in France]
Uetz P, Hallermann J. The Reptile Database. 1995-2024. [accessed 2024 Aug 15]. http://www.reptile-database.org/
Waters PD, Patel HR, Ruiz-Herrera A, Álvarez-González L, Lister NC, Simakov O, Ezaz T, Kaur P, Frere C, Grutzner F, and et al. 2021. Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc. Natl. Acad. Sci. USA. 118(45). doi: 10.1073/pnas.2112494118.
Wood PL Jr, Grismer LL, Grismer JL, Neang T, Chav T. Holden J. 2010. A new cryptic species of Acanthosaura Gray, 1831 (Squamata: Agamidae) from Thailand and Cambodia. Zootaxa 2488, 22 – 38.
Yano CF, Bertollo LA, Ezaz T, Trifonov V, Sember A, Liehr T. 2017. Highly conserved Z and molecularly diverged W chromosomes in the fish genus Triportheus (Characiformes, Triportheidae). Heredity 118, 276–283. doi: 10.1038/hdy.2016.83
Young MJ, O’Meally D, Sarre SD, Georges A, Ezaz T. 2013. Molecular cytogenetic map of the central bearded dragon, Pogona vitticeps (Squamata: Agamidae). Chromosome Res. 21: 361–374. doi: 10.1007/s10577-013-9362-z.
Zongyun L, Mingqin G, Lu W, Ming C, Xiuqin W. 2004. Studies on spermary chromosomes and its meiosis of Japalura splendida Barbour and Dunn, 1919. Sichuan Dongwu. Sichuan J. Zool. 23(3):281-284.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Nattasuda Donbundit; Praween Supanuam; Sittisak Jantarat, Thaintip Kraiprom, Somsak Buathip, Sarun Jumrusthanasan, Sarawut Kaewsri, Phichaya Buasriyot, Weera Thongnetr, Sumalee Phimphan, Alongklod Tanomtong

This work is licensed under a Creative Commons Attribution 4.0 International License.
- Copyright on any open access article in a journal published byCaryologia is retained by the author(s).
- Authors grant Caryologia a license to publish the article and identify itself as the original publisher.
- Authors also grant any third party the right to use the article freely as long as its integrity is maintained and its original authors, citation details and publisher are identified.
- The Creative Commons Attribution License 4.0 formalizes these and other terms and conditions of publishing articles.
- In accordance with our Open Data policy, the Creative Commons CC0 1.0 Public Domain Dedication waiver applies to all published data in Caryologia open access articles.